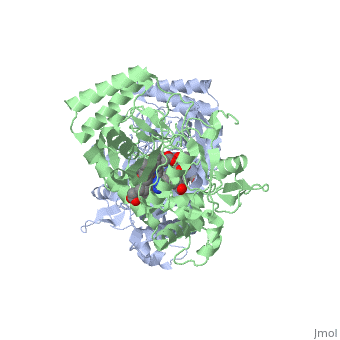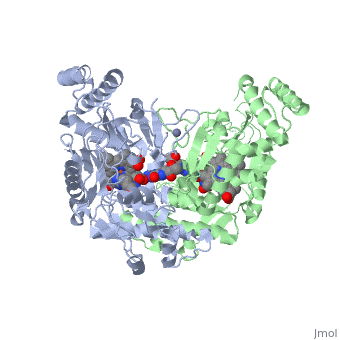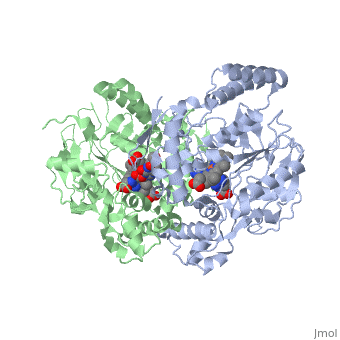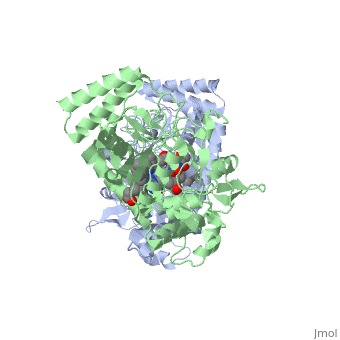
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Nitric Oxide Synthase
From Proteopedia
(Difference between revisions)
| (20 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='2g6h' size=' | + | <StructureSection load='2g6h' size='350' side='right' caption='Neuronal nitric oxide synthase dimer complex with cofactor tetrahydrobiopterin, acetate and Zn+2 (grey), (PDB entry [[2g6h]])' scene=''> |
| - | '''Nitric Oxide Synthase''' (NOS) is an enzyme catalysing the formation of L-citrulline and [http://en.wikipedia.org/wiki/Nitric_Oxide/ nitric oxide] (NO) from L-arginine. NOS is a homodimeric protein with 125- to 160-kDa per monomer. In mammals, NOS appears as 3 isozymes: neuronal NOS (nNOS) (for details see [[Nos1]]), cytokine-inducible NOS (iNOS) and endothelial NOS (eNOS). The N-terminal domain of NOS is an oxygenase domain (OD). NOS cofactors are: NADPH, FAD, FMN, heme and O2. See also [[Nos1]]. | + | '''Nitric Oxide Synthase''' (NOS) or '''Nitric Oxide Synthase oxygenase''' is an enzyme catalysing the formation of L-citrulline and [http://en.wikipedia.org/wiki/Nitric_Oxide/ nitric oxide] (NO) from L-arginine. NOS is a homodimeric protein with 125- to 160-kDa per monomer. In mammals, NOS appears as 3 isozymes: neuronal NOS (nNOS) (for details see [[Nos1]]), cytokine-inducible NOS (iNOS) and endothelial NOS (eNOS). The N-terminal domain of NOS is an '''oxygenase''' domain (OD). NOS cofactors are: NADPH, FAD, FMN, heme and O2. See also [[Nos1]]. |
=Introduction= | =Introduction= | ||
| Line 7: | Line 7: | ||
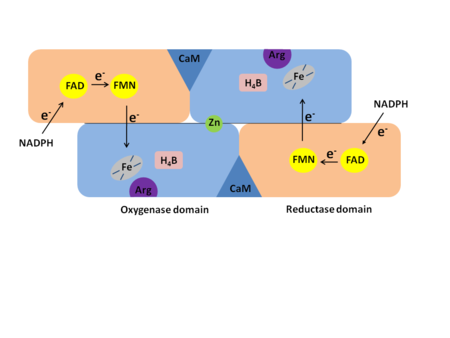
In mammals three isozymes of NOS have been identified: Neuronal NOS (nNOS), inducible NOS (iNOS), and endothelial NOS (eNOS)<ref>Flemming, I., Chapter 3, Biology of Nitric Oxide Synthases, from Microcirculation, Editors Ronald F. Tuma, R. F., Durán, W. N., and Ley, K., 2nd edition, Academic Press, 2008</ref>. (The NOS enzymes are found in numeral organisms. Most facts used here are from the human NOS, but sites from different organisms are used). Neuronal NOS is producing NO in the nervous tissue in both the peripheral and the central nervous system. nNOS is functioning in cell signaling and communication - a vital part of the nervous tissue. Inducible NOS is connected with the immune system. Endothelial NOS is controlling the amount of NO signaling in the endothelial cells eg. blood vessel dilation. An overview of the structural organization of the NOS homodimer is given below. All cofactors are included and the electron transfer pathway which takes place in NOS is indicated. | In mammals three isozymes of NOS have been identified: Neuronal NOS (nNOS), inducible NOS (iNOS), and endothelial NOS (eNOS)<ref>Flemming, I., Chapter 3, Biology of Nitric Oxide Synthases, from Microcirculation, Editors Ronald F. Tuma, R. F., Durán, W. N., and Ley, K., 2nd edition, Academic Press, 2008</ref>. (The NOS enzymes are found in numeral organisms. Most facts used here are from the human NOS, but sites from different organisms are used). Neuronal NOS is producing NO in the nervous tissue in both the peripheral and the central nervous system. nNOS is functioning in cell signaling and communication - a vital part of the nervous tissue. Inducible NOS is connected with the immune system. Endothelial NOS is controlling the amount of NO signaling in the endothelial cells eg. blood vessel dilation. An overview of the structural organization of the NOS homodimer is given below. All cofactors are included and the electron transfer pathway which takes place in NOS is indicated. | ||
{{Clear}} | {{Clear}} | ||
| - | [[Image:schematic_NOS.png | + | [[Image:schematic_NOS.png|left|thumb|450px]] |
{{Clear}} | {{Clear}} | ||
Each NOS monomer is composed of two domains: an N-terminal oxygenase domain and a C-terminal reductase domain. Each subunit is held together by a zinc ion, which is bound by two cysteines from each oxygenase domain. Calmodulin binds to a linker region between the two domains and is necessary for activity. The reductase domain supplies electrons for the NOS reaction which takes place in the oxygenase domain. The reductase domain contains two redox-active prosthetic groups, flavin adenine dinucleotide (FAD) and Flavin mononucleotide (FMN). Nicotinamide adenine dinucleotide phosphate(NADPH) binds to the domain and passes on an electron to FAD which passes the electron on to FMN. FMN passes the electron on to the heme in the oxygenase domain on the opposite subunit. The oxygenase domain contains 5,6,7,8-tetrahydrobiopterin (H<sub>4</sub>B) and the already mentioned heme. These two are also redox active groups. Besides Heme and H<sub>4</sub>B, the oxygenase domain binds the substrate L-arginine which takes part in the NO synthase reaction (see below). | Each NOS monomer is composed of two domains: an N-terminal oxygenase domain and a C-terminal reductase domain. Each subunit is held together by a zinc ion, which is bound by two cysteines from each oxygenase domain. Calmodulin binds to a linker region between the two domains and is necessary for activity. The reductase domain supplies electrons for the NOS reaction which takes place in the oxygenase domain. The reductase domain contains two redox-active prosthetic groups, flavin adenine dinucleotide (FAD) and Flavin mononucleotide (FMN). Nicotinamide adenine dinucleotide phosphate(NADPH) binds to the domain and passes on an electron to FAD which passes the electron on to FMN. FMN passes the electron on to the heme in the oxygenase domain on the opposite subunit. The oxygenase domain contains 5,6,7,8-tetrahydrobiopterin (H<sub>4</sub>B) and the already mentioned heme. These two are also redox active groups. Besides Heme and H<sub>4</sub>B, the oxygenase domain binds the substrate L-arginine which takes part in the NO synthase reaction (see below). | ||
| Line 19: | Line 19: | ||
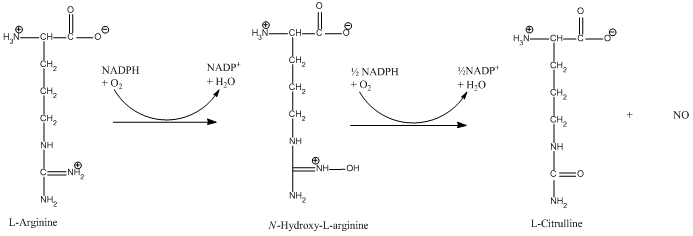
The NO production takes plase in the oxygenase domain, whereas the reductase domain provides the electrons necessary to drive the reaction in the oxygenase domain. | The NO production takes plase in the oxygenase domain, whereas the reductase domain provides the electrons necessary to drive the reaction in the oxygenase domain. | ||
The reaction is: | The reaction is: | ||
| - | [[image:Makroprojekt-chemdraw-1-.gif|frame|left|thumb| | + | [[image:Makroprojekt-chemdraw-1-.gif|frame|left|thumb|400px]] |
| + | {{Clear}} | ||
The reaction occurs in two steps. In the first step L-arginine is turned in to N-hydroxy-arginine. In the second step L-citrulline and NO are formed. The reaction utilizes 2O<sub>2</sub> and 3 electrons from 3/2 NADPH.<ref>PMID:17014963</ref> | The reaction occurs in two steps. In the first step L-arginine is turned in to N-hydroxy-arginine. In the second step L-citrulline and NO are formed. The reaction utilizes 2O<sub>2</sub> and 3 electrons from 3/2 NADPH.<ref>PMID:17014963</ref> | ||
| Line 45: | Line 46: | ||
===H<sub>4</sub>B=== | ===H<sub>4</sub>B=== | ||
[[image:bh4.png|left|frame|Structure of tetrahydrobiopterin]] | [[image:bh4.png|left|frame|Structure of tetrahydrobiopterin]] | ||
| - | + | {{Clear}} | |
<scene name='Sandbox_5/Nos_oxygenase_bh4/11'>H4B</scene> is an essential cofactor in NOS and in the [[aromatic amino acid hydroxylases]]. NOS contains two molecules of <scene name='Sandbox_5/Begge_h4b/1'>H4B</scene>, one in each monomer. The active site forms part of the cavity already described. This cavity can be visualized as a <scene name='Nitric_oxide_synthase/Substratebinding_distal_pocket/2'>tunnel</scene>. Here H<sub>4</sub>B helps substrate interactions by lining the active-center tunnel and hydrogen bonding to the heme propionate and to alfa helix 7. The propionate group and alpha helix 7 are also involved in the L-Arg binding. This gives H<sub>4</sub>B the opportunity to play an important role in the control of subunit interactions and active site formation. H<sub>4</sub>B is therefore more or less a structural cofactor and has a stabilizing effect. Its structural importance is further reconned to play a role in dimer formation (dimerization requires bound zinc ion along with H<sub>4</sub>B), and major conformational changes leading to the formation of the active site channelform<ref name="Raman">PMID:9875848</ref>. | <scene name='Sandbox_5/Nos_oxygenase_bh4/11'>H4B</scene> is an essential cofactor in NOS and in the [[aromatic amino acid hydroxylases]]. NOS contains two molecules of <scene name='Sandbox_5/Begge_h4b/1'>H4B</scene>, one in each monomer. The active site forms part of the cavity already described. This cavity can be visualized as a <scene name='Nitric_oxide_synthase/Substratebinding_distal_pocket/2'>tunnel</scene>. Here H<sub>4</sub>B helps substrate interactions by lining the active-center tunnel and hydrogen bonding to the heme propionate and to alfa helix 7. The propionate group and alpha helix 7 are also involved in the L-Arg binding. This gives H<sub>4</sub>B the opportunity to play an important role in the control of subunit interactions and active site formation. H<sub>4</sub>B is therefore more or less a structural cofactor and has a stabilizing effect. Its structural importance is further reconned to play a role in dimer formation (dimerization requires bound zinc ion along with H<sub>4</sub>B), and major conformational changes leading to the formation of the active site channelform<ref name="Raman">PMID:9875848</ref>. | ||
[[image:H4b_hydrogenbindinger2.png|thumb|left|Hydrogenbondings in H<sub>4</sub>B binding site ([[2nse]]) ]] | [[image:H4b_hydrogenbindinger2.png|thumb|left|Hydrogenbondings in H<sub>4</sub>B binding site ([[2nse]]) ]] | ||
| + | {{Clear}} | ||
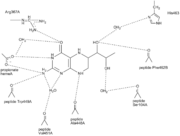
| + | The H<sub>4</sub>B is bound by H-bonds to several of the residues surrounding it. For example is O4 of H<sub>4</sub>B H-bonded to Arg 367 and H<sub>4</sub>B N3 is H-bonded to one of the heme propionate groups ( the heme propionate group has two carboxylate oxygens in use for H-bonds)<ref name="Raman"/>. The overall picture of all the H-bonds can be seen by clicking on the figure on the left. | ||
| - | The H<sub>4</sub>B is bound by H-bonds to several of the residues surrounding it. For example is O4 of H<sub>4</sub>B H-bonded to Arg 367 and H<sub>4</sub>B N3 is H-bonded to one of the heme propionate groups ( the heme propionate group has two carboxylate oxygens in use for H-bonds)<ref name="Raman"/>. The overall picture of all the H-bonds can be seen by clicking on the figure on the left. [[image:mette.png|thumb|right|model for NOS oxygen activation]] | ||
But H<sub>4</sub>B is not only a structural cofactor, it also plays a very important role in NO synthesis, donating an electron to the heme.<ref name="heme">PMID:12237227</ref> H<sub>4</sub>B can deliver an electron to the heme much faster than the reductase domain can, therefor H<sub>4</sub>B is used by NOS in the Arg hydroxylation, activating O<sub>2</sub> by providing the second electron. Thus, H<sub>4</sub>B is a kinetically prefered electron donor. As shown in the reaction (bottom right, click for enlargement) the second electron, that H<sub>4</sub>B donates, helps the Fe<sup>II</sup>O<sub>2</sub> intermediate to be reduced to oxidants that are able to react with Arg and N-hydroxy-L-arginine (NOHA) <ref name="heme"/> If H<sub>4</sub>B was not present the Fe<sup>II</sup>O<sub>2</sub> intermediate would decay to superoxide and ferric enzyme due to the reductase domain being slower to deliver an electron than the proces of decay is to happen. H<sub>4</sub>B is faster than both of these processes<ref name="heme"/>. | But H<sub>4</sub>B is not only a structural cofactor, it also plays a very important role in NO synthesis, donating an electron to the heme.<ref name="heme">PMID:12237227</ref> H<sub>4</sub>B can deliver an electron to the heme much faster than the reductase domain can, therefor H<sub>4</sub>B is used by NOS in the Arg hydroxylation, activating O<sub>2</sub> by providing the second electron. Thus, H<sub>4</sub>B is a kinetically prefered electron donor. As shown in the reaction (bottom right, click for enlargement) the second electron, that H<sub>4</sub>B donates, helps the Fe<sup>II</sup>O<sub>2</sub> intermediate to be reduced to oxidants that are able to react with Arg and N-hydroxy-L-arginine (NOHA) <ref name="heme"/> If H<sub>4</sub>B was not present the Fe<sup>II</sup>O<sub>2</sub> intermediate would decay to superoxide and ferric enzyme due to the reductase domain being slower to deliver an electron than the proces of decay is to happen. H<sub>4</sub>B is faster than both of these processes<ref name="heme"/>. | ||
| + | [[image:mette.png|thumb|left|model for NOS oxygen activation]] | ||
| + | {{Clear}} | ||
PDB structures used in the section above: [[3nos]], [[2g6h]] | PDB structures used in the section above: [[3nos]], [[2g6h]] | ||
| Line 88: | Line 92: | ||
Structures used in the section above: [[1tll]] | Structures used in the section above: [[1tll]] | ||
| - | </StructureSection> | ||
| - | == 3D Structures of Nitric Oxide Synthase == | ||
| - | + | = 3D Structures of Nitric Oxide Synthase = | |
| - | + | [[Nitric Oxide Synthase 3D structures]] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[2amo]] – BsNOS – ''Bacillus subtilis''<br /> | ||
| - | **[[2an0]], [[2an2]] – BsNOS (mutant)<br /> | ||
| - | **[[2fbz]], [[2fc2]], [[2fc1]] – BsNOS+NO<br /> | ||
| - | **[[1m7v]], [[1m7z]] – BsNOS+THF+arginine<br /> | ||
| - | **[[5g67]], [[4ugl]] – BsNOS + inhibitor<br /> | ||
| - | **[[2flq]] – NOS+arginine – ''Geobacillus stearothermophilus''<br /> | ||
| - | }} | ||
==References== | ==References== | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Flemming, I., Chapter 3, Biology of Nitric Oxide Synthases, from Microcirculation, Editors Ronald F. Tuma, R. F., Durán, W. N., and Ley, K., 2nd edition, Academic Press, 2008
- ↑ Gorren AC, Mayer B. Nitric-oxide synthase: a cytochrome P450 family foster child. Biochim Biophys Acta. 2007 Mar;1770(3):432-45. Epub 2006 Sep 1. PMID:17014963 doi:10.1016/j.bbagen.2006.08.019
- ↑ Fan B, Wang J, Stuehr DJ, Rousseau DL. NO synthase isozymes have distinct substrate binding sites. Biochemistry. 1997 Oct 21;36(42):12660-5. PMID:9376373 doi:http://dx.doi.org/10.1021/bi9715369
- ↑ Rousseau DL, Li D, Couture M, Yeh SR. Ligand-protein interactions in nitric oxide synthase. J Inorg Biochem. 2005 Jan;99(1):306-23. PMID:15598509 doi:http://dx.doi.org/10.1016/j.jinorgbio.2004.11.007
- ↑ 5.0 5.1 Raman CS, Li H, Martasek P, Kral V, Masters BS, Poulos TL. Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center. Cell. 1998 Dec 23;95(7):939-50. PMID:9875848
- ↑ 6.0 6.1 6.2 Wei CC, Wang ZQ, Meade AL, McDonald JF, Stuehr DJ. Why do nitric oxide synthases use tetrahydrobiopterin? J Inorg Biochem. 2002 Sep 20;91(4):618-24. PMID:12237227
- ↑ Li H, Igarashi J, Jamal J, Yang W, Poulos TL. Structural studies of constitutive nitric oxide synthases with diatomic ligands bound. J Biol Inorg Chem. 2006 Sep;11(6):753-68. Epub 2006 Jun 28. PMID:16804678 doi:http://dx.doi.org/10.1007/s00775-006-0123-8
- ↑ 8.0 8.1 Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, Dolphin E, Prongay AJ, Reichert P, Lundell DJ, Narula SK, Weber PC. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Biol. 1999 Mar;6(3):233-42. PMID:10074942 doi:http://dx.doi.org/10.1038/6675
- ↑ Crane BR, Rosenfeld RJ, Arvai AS, Ghosh DK, Ghosh S, Tainer JA, Stuehr DJ, Getzoff ED. N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization. EMBO J. 1999 Nov 15;18(22):6271-81. PMID:10562539 doi:http://dx.doi.org/10.1093/emboj/18.22.6271
- ↑ Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004 Sep 3;279(36):37918-27. Epub 2004 Jun 17. PMID:15208315 doi:http://dx.doi.org/10.1074/jbc.M406204200
Proteopedia Page Contributors and Editors (what is this?)
Sara Toftegaard Petersen, Michal Harel, Mathilde Thomsen, Michael Skovbo Windahl, Alexander Berchansky, Mette Trauelsen, Ann Taylor, Jaime Prilusky, Eran Hodis, Karl Oberholser