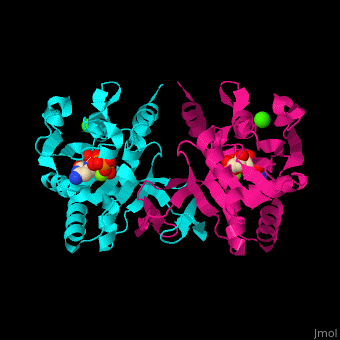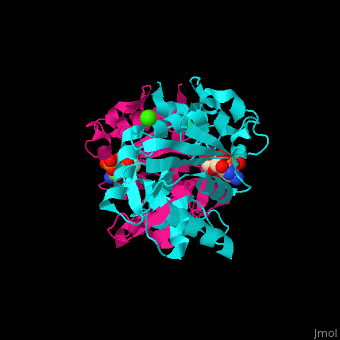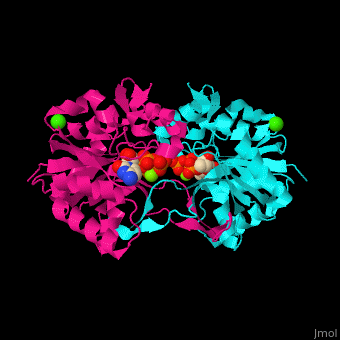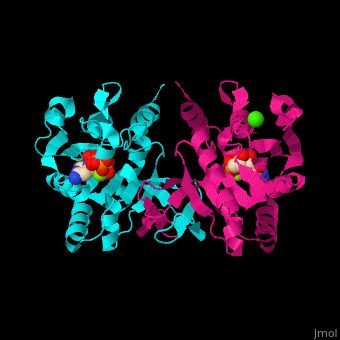MEP cytidylyltransferase
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
== Function == | == Function == | ||
| - | '''MEP cytidylyltransferase''' (MEPCT) catalyzes the conversion of 2C-methyl-D-erythritol 4-phosphate and CTP to 4-(cytidine 5’-diphospho)- 2C-methyl-D-erythritol (CDP-ME) and diphosphate. MEPCT participates in isoprenoid biosynthesis in many bacteria while eukaryotes use the mevalonic acid pathway for the isoprenoed biosynthesis<ref>PMID:11696548</ref>. | + | '''MEP cytidylyltransferase''' (MEPCT) catalyzes the conversion of 2C-methyl-D-erythritol 4-phosphate and CTP to 4-(cytidine 5’-diphospho)- 2C-methyl-D-erythritol (CDP-ME) and diphosphate. MEPCT participates in isoprenoid biosynthesis in many bacteria while eukaryotes use the mevalonic acid pathway for the isoprenoed biosynthesis<ref>PMID:11696548</ref>. '''IspD/IspDF''' is a bifunctional enzyme which catalyzes the formation of 4-diphosphocytidyl-2-C-methyl-D-erithritol 4-phophate and the conversion or CDP-ME2P to ME-CPP<ref>PMID:15233799</ref>. |
== Relevance == | == Relevance == | ||
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
| - | The biological assembly of ''E. coli'' MEPCT is <scene name='59/595219/Cv/ | + | The biological assembly of ''E. coli'' MEPCT is <scene name='59/595219/Cv/8'>homodimer</scene>. The <scene name='59/595219/Cv/9'>active site of MEPCT contains an Mg+2 ion</scene><ref>PMID:11427897</ref>. Water molecules shown as red spheres. <scene name='59/595219/Cv/10'>Whole active site</scene>. |
| - | + | ||
==3D structures of MEP cytidylyltransferase== | ==3D structures of MEP cytidylyltransferase== | ||
| + | [[MEP cytidylyltransferase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
Current revision
| |||||||||||
References
- ↑ Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, Madison EL. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J Biol Chem. 2002 Jan 18;277(3):2160-8. Epub 2001 Nov 5. PMID:11696548 doi:10.1074/jbc.M109830200
- ↑ Gabrielsen M, Rohdich F, Eisenreich W, Grawert T, Hecht S, Bacher A, Hunter WN. Biosynthesis of isoprenoids: a bifunctional IspDF enzyme from Campylobacter jejuni. Eur J Biochem. 2004 Jul;271(14):3028-35. doi: 10.1111/j.1432-1033.2004.04234.x. PMID:15233799 doi:http://dx.doi.org/10.1111/j.1432-1033.2004.04234.x
- ↑ Tsang A, Seidle H, Jawaid S, Zhou W, Smith C, Couch RD. Francisella tularensis 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase: kinetic characterization and phosphoregulation. PLoS One. 2011;6(6):e20884. doi: 10.1371/journal.pone.0020884. Epub 2011 Jun 9. PMID:21694781 doi:http://dx.doi.org/10.1371/journal.pone.0020884
- ↑ Richard SB, Bowman ME, Kwiatkowski W, Kang I, Chow C, Lillo AM, Cane DE, Noel JP. Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis. Nat Struct Biol. 2001 Jul;8(7):641-8. PMID:11427897 doi:10.1038/89691