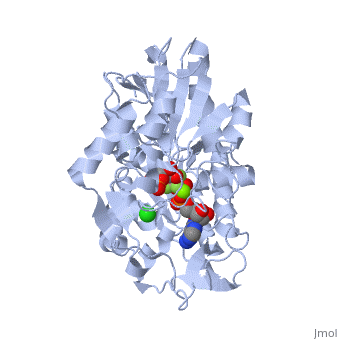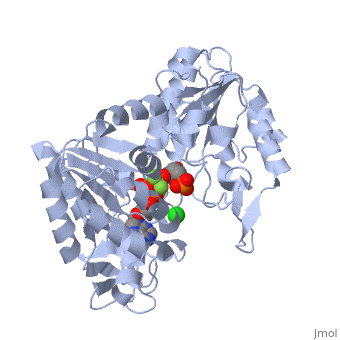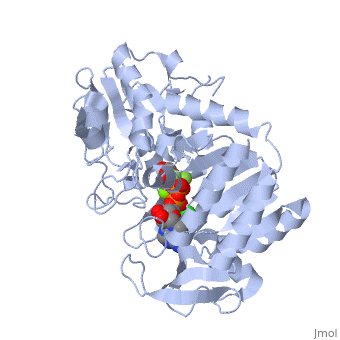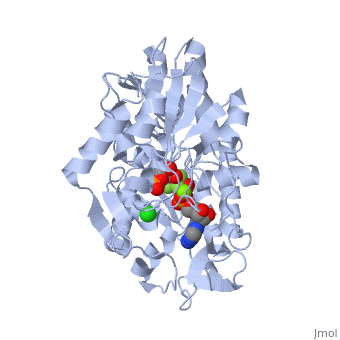
Phosphoglycerate Kinase
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
== PGK in the Glycolysis Cycle == | == PGK in the Glycolysis Cycle == | ||
| - | '''Phosphoglycerate kinase''' is a crucial enzyme in the glycolysis cycle. This cycle is a series of ten reactions which ultimately breaks down glucose into pyruvate while generating 2 NADH and 2 ATP molecules. Phosphoglycerate kinase is the seventh enzyme in the cycle which catalyzes the reaction of 1,3-Biphosphoglycerate and ADP to produce <scene name='Shane_Harmon_Sandbox/Product/2'>3-Phosphoglycerate</scene> and <scene name='Shane_Harmon_Sandbox/Atp/4'>ATP</scene>. This method for ATP production is known as substrate-level phosphorylation because it produces energy storing ATP molecules without the use of oxygen, NADH, or an ATPase. The reaction is highly exergonic allowing it to be coupled with the less thermodynamically favored GADPH reaction of the cycle so both reactions occur spontaneously. See [[Glycolysis Enzymes]]. | + | '''Phosphoglycerate kinase''' is a crucial enzyme in the glycolysis cycle. This cycle is a series of ten reactions which ultimately breaks down glucose into pyruvate while generating 2 NADH and 2 ATP molecules. Phosphoglycerate kinase is the seventh enzyme in the cycle which catalyzes the reaction of 1,3-Biphosphoglycerate and ADP to produce <scene name='Shane_Harmon_Sandbox/Product/2'>3-Phosphoglycerate</scene> and <scene name='Shane_Harmon_Sandbox/Atp/4'>ATP</scene>. This method for ATP production is known as substrate-level phosphorylation because it produces energy storing ATP molecules without the use of oxygen, NADH, or an ATPase. The reaction is highly exergonic allowing it to be coupled with the less thermodynamically favored GADPH reaction of the cycle so both reactions occur spontaneously. See [[Glycolysis Enzymes]], [[Gluconeogenesis]]. |
== Structure == | == Structure == | ||
| Line 9: | Line 9: | ||
PGK structure shows an open-to-close transition upon hinge bending. PG assumes the open conformation upon release of PGA and ATP. The closed conformation active site contains PGA, ADP and AlF<sub>4</sub>-1 ion which mimics the phosphate ion<ref>PMID:21549713</ref>. | PGK structure shows an open-to-close transition upon hinge bending. PG assumes the open conformation upon release of PGA and ATP. The closed conformation active site contains PGA, ADP and AlF<sub>4</sub>-1 ion which mimics the phosphate ion<ref>PMID:21549713</ref>. | ||
| - | *<scene name='38/387911/Cv/ | + | *<scene name='38/387911/Cv/10'>Phosphoglycerate binding site</scene>. |
| - | *<scene name='38/387911/Cv/ | + | *<scene name='38/387911/Cv/11'>AlF4- binding site</scene>. |
| - | *<scene name='38/387911/Cv/ | + | *<scene name='38/387911/Cv/12'>ADP binding site</scene>. |
| - | *<scene name='38/387911/Cv/ | + | *<scene name='38/387911/Cv/13'>Whole binding site</scene>. Water molecules are shown as red spheres. |
== Reaction Mechanism == | == Reaction Mechanism == | ||
| Line 28: | Line 28: | ||
== Kinetics == | == Kinetics == | ||
| - | Given that Phosphoglycerate kinase is a monomeric protein standard Michealis-Menton kinetics would be expected; however, this is not the case. Multiple experiments have shown that the data, when transformed into either double-reciprocal or Eadie-Hofstee plots is non-linear; Eadie-Hofstee plots curve upward. One possible explanation for the non-linearity, negative cooperativity, is ruled out because PGK does not have multiple subunits. In one study that conducted kinetic tests with a 1000 fold range of substrates, at the highest concentrations of substrate the rate was still increasing; this puts the Km value in the 2-5mM range <ref> | + | Given that Phosphoglycerate kinase is a monomeric protein standard Michealis-Menton kinetics would be expected; however, this is not the case. Multiple experiments have shown that the data, when transformed into either double-reciprocal or Eadie-Hofstee plots is non-linear; Eadie-Hofstee plots curve upward. One possible explanation for the non-linearity, negative cooperativity, is ruled out because PGK does not have multiple subunits. In one study that conducted kinetic tests with a 1000 fold range of substrates, at the highest concentrations of substrate the rate was still increasing; this puts the Km value in the 2-5mM range <ref>PMID:348474 </ref>. Recently, a new model was proposed to explain this conflict between the seemingly negative cooperative kinetics and the monomeric structure of PGK. The enzyme may form a complex with the metabolic enzyme glyceraldehyde-3-phosphate dehydrogenase. This multi-subunit complex would be capable of the negative cooperativity that is indicated by the non-linear kinetics of PGK<ref>PMID:16667700 </ref>. |
== Regulation == | == Regulation == | ||
| Line 37: | Line 37: | ||
Recent study of PGK has revolved around its function in tumor formation and growth. It has been shown that in addition to catalyzing its normal reaction of 1,3-Biphosphoglycerate and ADP to ATP and 3-Phosphoglycerate, PGK can also function to cleave disulfide bonds. Specifically, the review of sulfide bond cleavage indicates PGK has been shown to cleave disulfide bonds in the protein zymogen plasmin to produce the active form of the protein. The active form of plasmin is responsible for angiogenesis or blood vessel formation in tumors<ref>PMID:12189052</ref> Without the formation of blood vessels in tumors, nutrients are limited and tumor growth is therfore limited. Once blood vessels are established growth can rapidly increase. The fact that tumor cells secrete PGK to allow blood vessel formation through the activation of the zymogen plasmin has important implications for understanding its regulation. If the regulation of PGK in tumor cells can be understood, it might be possible to inhibit the overproduction and secretion of PGK to limit angiogenesis in tumors. | Recent study of PGK has revolved around its function in tumor formation and growth. It has been shown that in addition to catalyzing its normal reaction of 1,3-Biphosphoglycerate and ADP to ATP and 3-Phosphoglycerate, PGK can also function to cleave disulfide bonds. Specifically, the review of sulfide bond cleavage indicates PGK has been shown to cleave disulfide bonds in the protein zymogen plasmin to produce the active form of the protein. The active form of plasmin is responsible for angiogenesis or blood vessel formation in tumors<ref>PMID:12189052</ref> Without the formation of blood vessels in tumors, nutrients are limited and tumor growth is therfore limited. Once blood vessels are established growth can rapidly increase. The fact that tumor cells secrete PGK to allow blood vessel formation through the activation of the zymogen plasmin has important implications for understanding its regulation. If the regulation of PGK in tumor cells can be understood, it might be possible to inhibit the overproduction and secretion of PGK to limit angiogenesis in tumors. | ||
| - | </StructureSection> | ||
==3D structures of phosphoglycerate kinase == | ==3D structures of phosphoglycerate kinase == | ||
| + | [[Phosphoglycerate kinase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Additional Resources== | ==Additional Resources== | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ 1.0 1.1 1.2 1.3 Auerbach G, Huber R, Grattinger M, Zaiss K, Schurig H, Jaenicke R, Jacob U. Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability. Structure. 1997 Nov 15;5(11):1475-83. PMID:9384563
- ↑ Lallemand P, Chaloin L, Roy B, Barman T, Bowler MW, Lionne C. Interaction of human 3-phosphoglycerate kinase with its two substrates: is substrate antagonism a kinetic advantage? J Mol Biol. 2011 Jun 24;409(5):742-57. Epub 2011 Apr 27. PMID:21549713 doi:10.1016/j.jmb.2011.04.048
- ↑ Voet, Donald et al. 2008. Fundamentals of Biochemistry. 3rd ed. 499
- ↑ Blake CC, Rice DW. Phosphoglycerate kinase. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):93-104. PMID:6115427
- ↑ Vas M, Varga A, Graczer E. Insight into the Mechanism of Domain Movements and their Role in Enzyme Function: Example of 3-Phosphoglycerate Kinase. Curr Protein Pept Sci. 2010 Jan 21. PMID:20088776
- ↑ Haran G, Haas E, Szpikowska BK, Mas MT. Domain motions in phosphoglycerate kinase: determination of interdomain distance distributions by site-specific labeling and time-resolved fluorescence energy transfer. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11764-8. PMID:1465395
- ↑ Scopes RK. The steady-state kinetics of yeast phosphoglycerate kinase. Anomalous kinetic plots and the effects of salts on activity. Eur J Biochem. 1978 Apr 17;85(2):503-16. PMID:348474
- ↑ Macioszek J, Anderson JB, Anderson LE. Isolation of chloroplastic phosphoglycerate kinase : kinetics of the two-enzyme phosphoglycerate kinase/glyceraldehyde-3-phosphate dehydrogenase couple. Plant Physiol. 1990 Sep;94(1):291-6. PMID:16667700
- ↑ Wu S, Storey JM, Storey KB. Phosphoglycerate kinase 1 expression responds to freezing, anoxia, and dehydration stresses in the freeze tolerant wood frog, Rana sylvatica. J Exp Zool A Ecol Genet Physiol. 2009 Jan 1;311(1):57-67. doi: 10.1002/jez.495. PMID:18785212 doi:http://dx.doi.org/10.1002/jez.495
- ↑ Hogg PJ. Biological regulation through protein disulfide bond cleavage. Redox Rep. 2002;7(2):71-7. doi: 10.1179/135100002125000299. PMID:12189052 doi:http://dx.doi.org/10.1179/135100002125000299
Proteopedia Page Contributors and Editors (what is this?)
Shane Harmon, Michal Harel, Joel L. Sussman, Brandon Tritle, David Canner, Alexander Berchansky