This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Green Fluorescent Protein
From Proteopedia
| (19 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
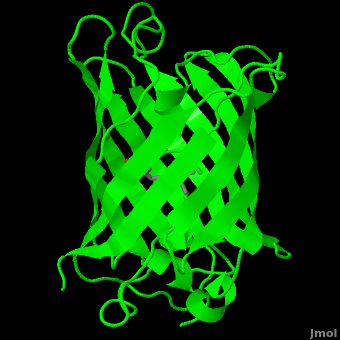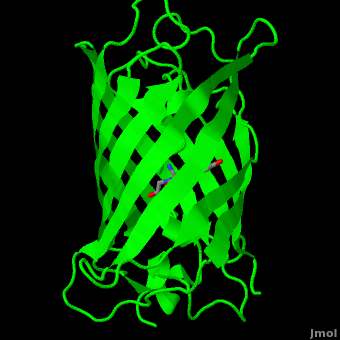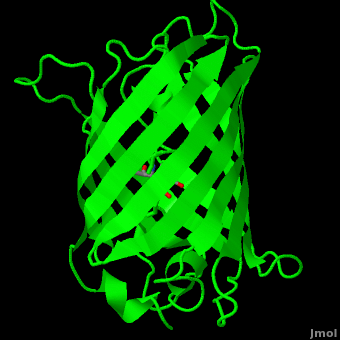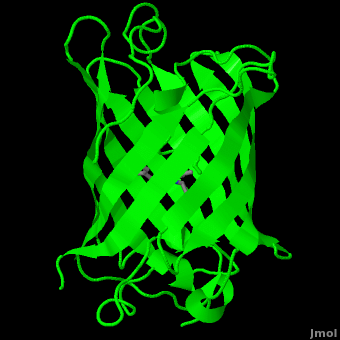
<StructureSection load='1ema' size='350' side='right' scene='Green_Fluorescent_Protein/1ema_gfp_default/2' caption='Green fluorescent protein complex with peptide-derived chromophore ([[1ema]])' > | <StructureSection load='1ema' size='350' side='right' scene='Green_Fluorescent_Protein/1ema_gfp_default/2' caption='Green fluorescent protein complex with peptide-derived chromophore ([[1ema]])' > | ||
==Function== | ==Function== | ||
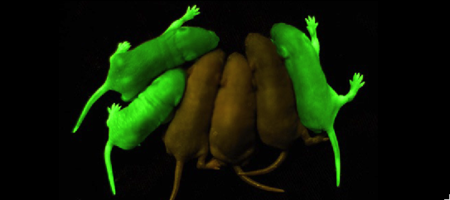
| - | '''Green fluorescent protein (GFP)''' is a | + | '''Green fluorescent protein (GFP)''' is a bioluminescent polypeptide consisting of 238 residues isolated from the body of ''Aequorea victoria'' jellyfish.<ref name="PDBsum">[http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=main.html], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.</ref> GFP converts the blue chemiluminescent of aequorin in the jellyfish into green fluorescent light.<ref name="Yang">[http://www-bioc.rice.edu/Bioch/Phillips/Papers/gfpbio.html], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.</ref> It remains unclear why these jellyfish use fluorescence, why green is better than blue, or why they produce a separate protein for green fluorescence as opposed to simply mutating the present aequorin to shift its wavelength,<ref name="Tsien" /> but in the laboratory, GFP can be incorporated into a variety of biological systems in order to function as a marker protein. Since its discovery in 1962, GFP has come to play a significant role in research as a tool to monitor gene expression, cellular localization, protein mobility, intracellular trafficking, and interactions between various membrane and cytoplasmic proteins, as well as many others. '''Superfolder GFP''' does not misfold when fused to other proteins. |
| + | |||
| + | See also<br /> | ||
[[Green Fluorescent Protein: Research Tool]].<ref name="Haldar"> [http://www.springerlink.com/content/wvg513864266g77n/fulltext.pdf], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on [http://en.wikipedia.org/wiki/Douglas_Prasher Douglas Prasher], [http://en.wikipedia.org/wiki/Martin_Chalfie Martin Chalfie] and [http://en.wikipedia.org/wiki/Roger_Tsien Roger Tsien].</ref><br /> | [[Green Fluorescent Protein: Research Tool]].<ref name="Haldar"> [http://www.springerlink.com/content/wvg513864266g77n/fulltext.pdf], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on [http://en.wikipedia.org/wiki/Douglas_Prasher Douglas Prasher], [http://en.wikipedia.org/wiki/Martin_Chalfie Martin Chalfie] and [http://en.wikipedia.org/wiki/Roger_Tsien Roger Tsien].</ref><br /> | ||
| - | [[Colored & Bioluminescent Protein]]<br />In Hebrew: [[GFP (Hebrew)]] and [[Gfp vc2]]. | + | [[Colored & Bioluminescent Protein]]<br /> |
| + | For''' red fluorescent protein''' see [[MCherry Fluorescent Protein]].<br /> | ||
| + | In Hebrew: [[GFP (Hebrew)]] and [[Gfp vc2]]. | ||
==History== | ==History== | ||
| Line 20: | Line 24: | ||
Green fluorescent protein (<scene name='Green_Fluorescent_Protein/1ema_gfp_default/2'>default scene</scene>) is a 21 kDa protein consisting of 238 residues strung together<ref>Primary structure at [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=protein.html&r=wiring&l=1&chain=A www.ebi.aci.uk].</ref> to form a | Green fluorescent protein (<scene name='Green_Fluorescent_Protein/1ema_gfp_default/2'>default scene</scene>) is a 21 kDa protein consisting of 238 residues strung together<ref>Primary structure at [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=protein.html&r=wiring&l=1&chain=A www.ebi.aci.uk].</ref> to form a | ||
| - | <scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2'>secondary structure</scene> of five α-helices and one eleven-stranded β-pleated sheet,<ref name="PDBsum" /> where each strand contains nine to thirteen residues each.<ref name="Ormo" /> (To view the primary and secondary structure of GFP, go to .) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.<ref name="Phillips">PMID: 9434902</ref> This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.<ref name="Yang" /> In GFP, the structure is so regular that <scene name='Green_Fluorescent_Protein/Water_stripes/1'>"stripes"</scene> of water molecules (red) can be seen following the structure of the barrel.<ref name="Phillips" /> Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,<ref name="Ormo" /> creating what is referred to as a “β-can” formation.<ref name="Phillips" /> The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.<ref name="Phillips" /> Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.<ref name="Yang" /> | + | <scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2'>secondary structure</scene> of five α-helices and one eleven-stranded β-pleated sheet,<ref name="PDBsum" /> where each strand contains nine to thirteen residues each.<ref name="Ormo" /> (To view the primary and secondary structure of GFP, go to https://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=1EMA.) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.<ref name="Phillips">PMID: 9434902</ref> This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.<ref name="Yang" /> In GFP, the structure is so regular that <scene name='Green_Fluorescent_Protein/Water_stripes/1'>"stripes"</scene> of water molecules (red) can be seen following the structure of the barrel.<ref name="Phillips" /> Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,<ref name="Ormo" /> creating what is referred to as a “β-can” formation.<ref name="Phillips" /> The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.<ref name="Phillips" /> Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.<ref name="Yang" /> |
| + | |||
| + | One <jmol><jmolLink><script>script "/scripts/Green_Fluorescent_Protein/Central_helix/1.spt"; ppdiaCaptionCmd = "changeCaption('The central helix (shown in red) contains the fluorophore and runs through the barrel (shown as white transparent strands) along its axis (PDB-ID [[1ema]]). ','white','black');";javascript @ppdiaCaptionCmd;model 2;</script><text>α-helix</text></jmolLink></jmol> can be found running through the central axis of the β-barrel,<ref name="Haldar" /> roughly <scene name='Green_Fluorescent_Protein/Perpendicular/1'>perpendicular</scene> to the symmetry axis of the barrel.<ref name="Ormo">Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the ''Aequorea victoria'' green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.</ref> This helix is extremely important as it contains the fluorophore responsible for fluorescence.<ref name="Yang" /><ref name="Haldar" /> | ||
| + | |||
| + | The fluorophore is part of the polypeptide chain (i.e. covalently connencted). If you press the buttom below, it will show the connection. | ||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>select (64.C or 66.N1 or 66.C3 or 68.N); connect 1.1 1.6; select 64, 66, 68; wireframe 0.2; </script> | ||
| + | <text>make bonds</text> | ||
| + | </jmolButton> | ||
| + | </jmol> | ||
| + | |||
| + | This α-helix in particular is highly stabilized by the many <scene name='Green_Fluorescent_Protein/Spacefill/1'>contacts</scene> that are made with each strand of the barrel.<ref name="Andrews">PMID:18713871</ref> | ||
| + | |||
| - | One <scene name='Green_Fluorescent_Protein/Central_helix/1'>α-helix</scene> can be found running through the central axis of the β-barrel,<ref name="Haldar" /> roughly <scene name='Green_Fluorescent_Protein/Perpendicular/1'>perpendicular</scene> to the symmetry axis of the barrel.<ref name="Ormo">Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the ''Aequorea victoria'' green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.</ref> This helix is extremely important as it contains the fluorophore responsible for fluorescence.<ref name="Yang" /><ref name="Haldar" /> This α-helix in particular is highly stabilized by the many <scene name='Green_Fluorescent_Protein/Spacefill/1'>contacts</scene> that are made with each strand of the barrel.<ref name="Andrews">PMID:18713871</ref> | ||
{{Link Toggle FancyCartoonHighQualityView}}. | {{Link Toggle FancyCartoonHighQualityView}}. | ||
| Line 32: | Line 48: | ||
The <scene name='10/100139/Chromophore/2'>chromophore</scene> (<scene name='10/100139/Green_fluorescent_protein/1'>top view</scene>) of GFP is located at the center of the β-barrel with a wild-type excitation peak of 395 nm, and a minor peak at 475 nm (about three times less intense<ref name="Tsien" />) <ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> with extinction coefficients of approximately 30,000 and 7,000 M<sup>-1</sup> cm<sup>-1</sup>, respectively.<ref name="Yang" /><ref name="Phillips" /> Interestingly, the ''Aequorea victoria'' jellyfish utilizes the smaller of the two excitation peaks as pure aequorin emits a light of 470 nm.<ref name="Tsien">Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.</ref> The relative amplitudes of these two excitation peaks can vary depending on environmental factors and previous illumination.<ref name="Ormo" /> For example, continued excitation leads to a diminution of the 395 nm excitation peak with a reciprocal amplification of the 475 nm peak.<ref name="Phillips" /> Regardless of absorption, the chromophore of GFP emits light of 508 nm.<ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> | The <scene name='10/100139/Chromophore/2'>chromophore</scene> (<scene name='10/100139/Green_fluorescent_protein/1'>top view</scene>) of GFP is located at the center of the β-barrel with a wild-type excitation peak of 395 nm, and a minor peak at 475 nm (about three times less intense<ref name="Tsien" />) <ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> with extinction coefficients of approximately 30,000 and 7,000 M<sup>-1</sup> cm<sup>-1</sup>, respectively.<ref name="Yang" /><ref name="Phillips" /> Interestingly, the ''Aequorea victoria'' jellyfish utilizes the smaller of the two excitation peaks as pure aequorin emits a light of 470 nm.<ref name="Tsien">Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.</ref> The relative amplitudes of these two excitation peaks can vary depending on environmental factors and previous illumination.<ref name="Ormo" /> For example, continued excitation leads to a diminution of the 395 nm excitation peak with a reciprocal amplification of the 475 nm peak.<ref name="Phillips" /> Regardless of absorption, the chromophore of GFP emits light of 508 nm.<ref name="Yang" /><ref name="Cubitt" /><ref name="Ormo" /><ref name="Phillips" /> | ||
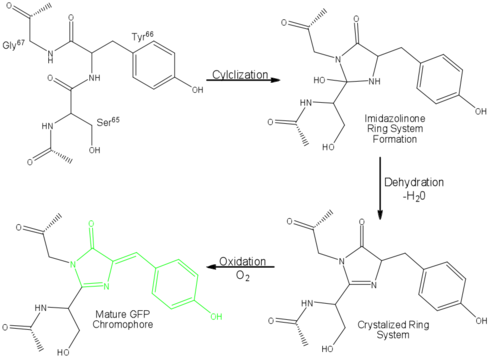
| - | Three amino residues in the central α-helix constitute the fluorophore of GFP: Ser<sup>65</sup>Tyr<sup>66</sup>Gly<sup>67</sup> (see below). Tsien et al. discovered that this tri-peptide sequence is post-translationally modified by internal cyclization and oxidation<ref name="Haldar" /> to produce a <scene name=' | + | Three amino residues in the central α-helix constitute the fluorophore of GFP: Ser<sup>65</sup>Tyr<sup>66</sup>Gly<sup>67</sup> (see below) or of EGFP: of GFP: Thr<sup>65</sup>Tyr<sup>66</sup>Gly<sup>67</sup>. Tsien et al. discovered that this tri-peptide sequence is post-translationally modified by internal cyclization and oxidation<ref name="Haldar" /> to produce a <scene name='10/100139/Chromophore_structure/3'>4-(p-hydroxybenzylidene)-imidazolidin-5-one</scene> structure (highlight atoms from <jmol><jmolLink><script> select (*.C1, *.CA1, *.N1, *.CB1, *.CG1, *.OG1) and 66; selectionHalos ON; delay 1.5;selectionHalos OFF;</script><text>⚞Thr 65⚟</text></jmolLink> </jmol>, <jmol><jmolLink><script> select (*.C2, *.O2, *.CA2, *.N2, *.CB2, *.CG2, *.CD1, *.CD2, *.CE1, *.CE2, *.CZ, *.OH) and 66; selectionHalos ON; delay 1.5;selectionHalos OFF;</script><text>⚞Tyr 66⚟</text></jmolLink> </jmol>, <jmol><jmolLink><script> select (*.O3,*.C3,*.CA3,*.N3) and 66; selectionHalos ON; delay 1.5;selectionHalos OFF;</script><text>⚞Gly 67⚟</text></jmolLink></jmol>).<ref name="Yang" /> Studies with E. coli proposed a sequential mechanism for the formation of the fluorophore that was initiated by a rapid cyclization between Ser<sup>65</sup> and Gly<sup>67</sup> to form an imidazolin-5-one intermediate.<ref name="Yang" /> This rapid cyclization is carried out via nucleophilic attack of the amino group from Gly<sup>67</sup> on the carbonyl group of Ser<sup>65</sup> to form a five-membered ring. The loss of water then forms the imidazolin-5-one intermediate.<ref name="Cubitt" /> Cyclization is succeeded by a much slower rate-limiting oxygenation of the Tyr<sup>66</sup> hydroxybenzyl side chain by atmospheric oxygen (No fluorescence was seen in anaerobically grown E. coli.), resulting in the 4-(p-hydroxybenzylidene)-imidazolidin-5-one stucture.<ref name="Yang" /><ref name="Cubitt" /><ref name="Phillips" /> The double bond that results from this series of reactions results in the linkage of the two π-systems of the rings, forming a <scene name='10/100139/Chromophore/5'>larger conjugated system</scene> essential for fluorophore stability. <ref name="Bublitz"> Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.</ref> |
[[Image:GFP Chromophore.png|center|489x360px]] | [[Image:GFP Chromophore.png|center|489x360px]] | ||
The process is completely auto-catalytic such that there are no known co-factors or enzymatic components required.<ref name="Yang" /> Despite the stability of the final product, while the chromophore is forming, the environmental temperature cannot drop below 30°C or the yield of viable GFP will decrease substantially.<ref name="Yang" /><ref name="Phillips" /> This, of course, is not an issue for the protein in nature as the jellyfish is unlikely to encounter waters of this degree in the Pacific Northwest.<ref name="Tsien" /> Such a temperature sensitivity is only relevant during formation as the stability of the final product is maintained through a network of close contacts surrounding the fluorophore.<ref name="Yang" /> This, however, can and has been used in [[pulse-chase experiments]] in which the GFP-expressing cells are exposed to varying temperatures in place of labeled vs. unlabeled trials.<ref name="Tsien" /> | The process is completely auto-catalytic such that there are no known co-factors or enzymatic components required.<ref name="Yang" /> Despite the stability of the final product, while the chromophore is forming, the environmental temperature cannot drop below 30°C or the yield of viable GFP will decrease substantially.<ref name="Yang" /><ref name="Phillips" /> This, of course, is not an issue for the protein in nature as the jellyfish is unlikely to encounter waters of this degree in the Pacific Northwest.<ref name="Tsien" /> Such a temperature sensitivity is only relevant during formation as the stability of the final product is maintained through a network of close contacts surrounding the fluorophore.<ref name="Yang" /> This, however, can and has been used in [[pulse-chase experiments]] in which the GFP-expressing cells are exposed to varying temperatures in place of labeled vs. unlabeled trials.<ref name="Tsien" /> | ||
| - | As the central α-helix is not located directly in the center of the β-barrel, cavities of differing area exist on either side of the chromophore. The larger cavity, consisting of about 135 Å,<ref name="Ormo" /> does not open out to the bulk solvent, but rather houses <scene name='Green_Fluorescent_Protein/Water_molecules/1'>four water molecules</scene>.<ref name="Ormo" /><ref name="Van">van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.</ref> Had this space not been occupied, it would be expected to considerably destabilize the protein as a whole. The hydrogen bonding created by the presence of the water molecules, however, helps to link the buried <scene name='Green_Fluorescent_Protein/Gln69_glu222/1'>side chains</scene> of Glu<sup>222</sup> and Gln<sup>69</sup> that would otherwise be actively polar.<ref name="Ormo" /> Therefore, the water molecules are extremely important in establishing a hydrogen bonding network about the chromophor.<ref name="Lammich">PMID: 17040991</ref> | + | As the central α-helix is not located directly in the center of the β-barrel, cavities of differing area exist on either side of the chromophore. The larger cavity, consisting of about 135 cubic Å,<ref name="Ormo" /> does not open out to the bulk solvent, but rather houses <scene name='Green_Fluorescent_Protein/Water_molecules/1'>four water molecules</scene>.<ref name="Ormo" /><ref name="Van">van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.</ref> Had this space not been occupied, it would be expected to considerably destabilize the protein as a whole. The hydrogen bonding created by the presence of the water molecules, however, helps to link the buried <scene name='Green_Fluorescent_Protein/Gln69_glu222/1'>side chains</scene> of Glu<sup>222</sup> and Gln<sup>69</sup> that would otherwise be actively polar.<ref name="Ormo" /> Therefore, the water molecules are extremely important in establishing a hydrogen bonding network about the chromophor.<ref name="Lammich">PMID: 17040991</ref> |
The opposite side of the chromophore, however, is within close proximity of several aromatic and polar side chains. Several <scene name='Green_Fluorescent_Protein/Polar_interactions/2'>polar interactions</scene> between the surrounding residues and the chromophore are present including: hydrogen bonds of His<sup>148</sup>, Thr<sup>203</sup>, and Ser<sup>205</sup> with the phenolic hydroxyl of Tyr<sup>66</sup>; Arg<sup>96</sup> and Gln<sup>94</sup> with the carbonyl of the imidazolidinone ring; and hydrogen bonds of Glu<sup>222</sup> with the side chain of Thr<sup>65</sup>. Additional hydrogen bonding in the area around the chromophore helps to stabilize Arg<sup>96</sup> in the protonated form, which suggests the presence of a partial negative charge on the carbonyl oxygen of the imidazolidinone ring in the deprotonated fluorophore.<ref name="Ormo" /> Arg<sup>96</sup> and Gln<sup>94</sup> in turn help to steady the imidazolidone.<ref name="Yang" /> Therefore, it is thought that Arg<sup>96</sup> is essential for the formation of the fluorophore by catalyzing the initial ring closure.<ref name="Ormo" /> Tyr<sup>145</sup> provides a stabilizing <scene name='Green_Fluorescent_Protein/Edge_face_interaction/1'>edge-face interaction</scene><ref> [http://www.tim.hi-ho.ne.jp/dionisio/ Information about edge-face (CH/π) interactions].</ref> with the benzyl ring of the chromophore.<ref name="Ormo" /> The stability provided by the internal polar interactions are further augmented by the surrounding β-barrel. | The opposite side of the chromophore, however, is within close proximity of several aromatic and polar side chains. Several <scene name='Green_Fluorescent_Protein/Polar_interactions/2'>polar interactions</scene> between the surrounding residues and the chromophore are present including: hydrogen bonds of His<sup>148</sup>, Thr<sup>203</sup>, and Ser<sup>205</sup> with the phenolic hydroxyl of Tyr<sup>66</sup>; Arg<sup>96</sup> and Gln<sup>94</sup> with the carbonyl of the imidazolidinone ring; and hydrogen bonds of Glu<sup>222</sup> with the side chain of Thr<sup>65</sup>. Additional hydrogen bonding in the area around the chromophore helps to stabilize Arg<sup>96</sup> in the protonated form, which suggests the presence of a partial negative charge on the carbonyl oxygen of the imidazolidinone ring in the deprotonated fluorophore.<ref name="Ormo" /> Arg<sup>96</sup> and Gln<sup>94</sup> in turn help to steady the imidazolidone.<ref name="Yang" /> Therefore, it is thought that Arg<sup>96</sup> is essential for the formation of the fluorophore by catalyzing the initial ring closure.<ref name="Ormo" /> Tyr<sup>145</sup> provides a stabilizing <scene name='Green_Fluorescent_Protein/Edge_face_interaction/1'>edge-face interaction</scene><ref> [http://www.tim.hi-ho.ne.jp/dionisio/ Information about edge-face (CH/π) interactions].</ref> with the benzyl ring of the chromophore.<ref name="Ormo" /> The stability provided by the internal polar interactions are further augmented by the surrounding β-barrel. | ||
| Line 61: | Line 77: | ||
{{Link Toggle FancyCartoonHighQualityView}}. | {{Link Toggle FancyCartoonHighQualityView}}. | ||
| - | |||
| - | |||
| - | |||
== Using GFP as a Research Tool == | == Using GFP as a Research Tool == | ||
A description of some of the ways GFP is being used as a tool in research is at [[Green_Fluorescent_Protein:_Research_Tool]]. | A description of some of the ways GFP is being used as a tool in research is at [[Green_Fluorescent_Protein:_Research_Tool]]. | ||
| - | |||
| - | </StructureSection> | ||
| - | |||
| - | |||
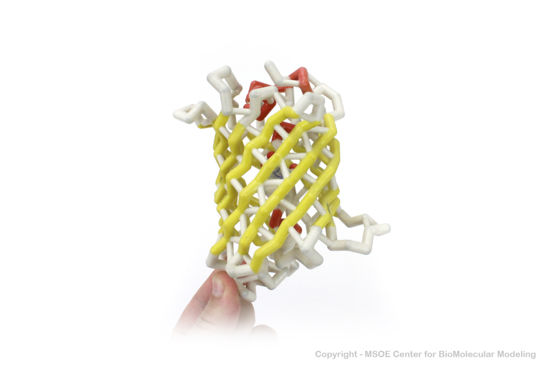
==3D Printed Physical Model of Green Fluorescent Protein (GFP)== | ==3D Printed Physical Model of Green Fluorescent Protein (GFP)== | ||
| Line 87: | Line 96: | ||
| - | ==3D structures of Green Fluorescent Protein (Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}}) == | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | + | ==3D structures of Green Fluorescent Protein == | |
| + | [[Green Fluorescent Protein 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Reference for this Structure== | ==Reference for this Structure== | ||
Current revision
| |||||||||||
Reference for this Structure
Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
References
- ↑ 1.0 1.1 [1], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 [2], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 [3], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on Douglas Prasher, Martin Chalfie and Roger Tsien.
- ↑ 5.0 5.1 5.2 5.3 [4], Shimomura O. The discovery of green fluorescent protein. Nobel Prize Lecture; 2009;; biographical background at Wikipedia.
- ↑ [5],Cowles D, Cowles J. Aequorea victoria. 2007. Walla Wall University.
- ↑ Primary structure at www.ebi.aci.uk.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 Phillips GN Jr. Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol. 1997 Dec;7(6):821-7. PMID:9434902
- ↑ Andrews BT, Gosavi S, Finke JM, Onuchic JN, Jennings PA. The dual-basin landscape in GFP folding. Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12283-8. Epub 2008 Aug 19. PMID:18713871
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 [6],Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien R. 1995. Understanding, improving, and using green fluorescent protein. Trends in Biochemical Sciences. 20(11): 448-455. DOI 0.1016/S0968-0004(00)89099-4.
- ↑ Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.
- ↑ van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.
- ↑ 14.0 14.1 Lammich L, Petersen MA, Nielsen MB, Andersen LH. The gas-phase absorption spectrum of a neutral GFP model chromophore. Biophys J. 2007 Jan 1;92(1):201-7. Epub 2006 Oct 13. PMID:17040991 doi:10.1529/biophysj.106.093674
- ↑ Information about edge-face (CH/π) interactions.
- ↑ Fang C, Frontiera RR, Tran R, Mathies RA. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature. 2009 Nov 12;462(7270):200-4. PMID:19907490 doi:10.1038/nature08527
Additional Resources
- For additional information, see: Colored & Bioluminescent Proteins
- First Glance
- PDBsum: 1ema
- RCSB PDB 1ema
- OCA
- UniProt: P42212
- Scop: P42212
- CATH: 1emaA00
- Pfam: PF01353
- InterPro: IPR000786
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
- Inside green fluorescent protein - editor's summary that accompanied structural detail of GFP chromophore on the cover of Nature.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, Karsten Theis, Eran Hodis, Laura Carbone, Karl Oberholser, Mark Hoelzer, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Joseph M. Steinberger, David Canner