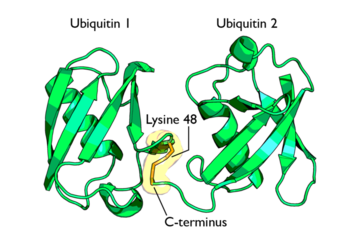Ubiquitin Structure & Function
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | ==Your Heading Here (maybe something like 'Structure')== | ||
<StructureSection load='1ubq' size='350' side='right' caption='Human ubiquitin, [[1ubq]])' scene=''> | <StructureSection load='1ubq' size='350' side='right' caption='Human ubiquitin, [[1ubq]])' scene=''> | ||
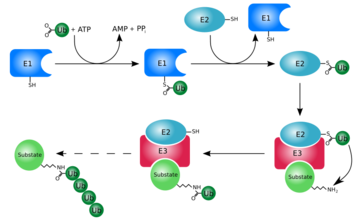
| - | [[Ubiquitin]] is a single 8565 M<sub>r</sub> polypeptide consisting of 76 amino acid residues | + | [[Ubiquitin]] is a single 8565 M<sub>r</sub> polypeptide consisting of 76 amino acid residues and is highly known for its role in ATP-dependent protein degradation<ref name="mainpaper">PMID: 3041007</ref> |
=Introduction= | =Introduction= | ||
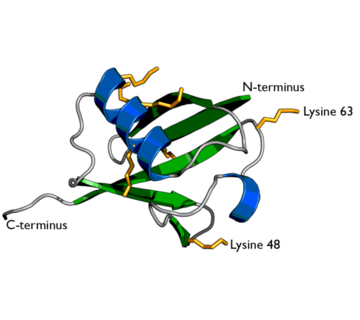
| - | Ubiquitin is one of the most highly conserved eukaryotic proteins. Primary structures found throughout ubiquitin are identical in all bovine, insects and human ubiquitin <ref name="mainpaper"/>. The only difference observed amongst these species is seen in the terminal Gly-Gly residues. Yeast and oat ubiquitin only differ in three of the 76 residues when compared to ubiquitin found in higher eukaryotes<ref name="mainpaper"/>.[[image:1ubiq.png| thumb |none | upright=2.0 |Ubiquitin structure: Arg74 in pink and Gly75 Gly76 in white.]] | + | Ubiquitin is one of the most highly conserved eukaryotic proteins. Primary structures found throughout ubiquitin are identical in all bovine, insects and human ubiquitin <ref name="mainpaper"/>. The only difference observed amongst these species is seen in the terminal Gly-Gly residues. Yeast and oat ubiquitin only differ in three of the 76 residues when compared to ubiquitin found in higher eukaryotes<ref name="mainpaper"/>. |
| + | [[image:1ubiq.png| thumb |none | upright=2.0 |Ubiquitin structure: Arg74 in pink and Gly75 Gly76 in white.]] | ||
| + | |||
| + | <!-- The content below was inserted by the ConSurf template --> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|right|200px]] | ||
| + | Check | ||
| + | <jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked> | ||
| + | select protein; define ~consurf_to_do selected; | ||
| + | consurf_initial_scene = true; | ||
| + | script /wiki/ConSurf/ub/1ubq_consurf.spt; | ||
| + | </scriptWhenChecked> | ||
| + | <scriptWhenUnchecked> | ||
| + | script /wiki/extensions/Proteopedia/spt/initialview01.spt; | ||
| + | </scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. | ||
| + | You may read the [[Conservation%2C_Evolutionary|explanation]] | ||
| + | of the method and the full data available from | ||
| + | [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ubq ConSurf]. | ||
| + | <!-- end of content inserted by the ConSurf template --> | ||
| + | |||
Ubiquitin can not only be found in the nucleus, but in the cytoplasm and cell-surface membrane as well<ref name="mainpaper"/>. | Ubiquitin can not only be found in the nucleus, but in the cytoplasm and cell-surface membrane as well<ref name="mainpaper"/>. | ||
| Line 39: | Line 62: | ||
Infectious agents can manipulate ubiquitin or deubiquitination and one such protein is Chlamydia trachomatis. Chlamydia trachomatis' protein Cdu-1 catalyzes the hydrolysis of ubiquitin chains from Mcl-1. When polyubiquitnated, Mcl-1 is destined to be degraded by the proteasome, lowering the level of Mcl-1 and subsequently leading to apoptosis. The activity of Cdu-1 counteracts this by removing the ubiquitin, thus leading to higher levels of Mcl-1 in the cell. Additional information can be found here [[User:Karsten Theis/5B5Q]] | Infectious agents can manipulate ubiquitin or deubiquitination and one such protein is Chlamydia trachomatis. Chlamydia trachomatis' protein Cdu-1 catalyzes the hydrolysis of ubiquitin chains from Mcl-1. When polyubiquitnated, Mcl-1 is destined to be degraded by the proteasome, lowering the level of Mcl-1 and subsequently leading to apoptosis. The activity of Cdu-1 counteracts this by removing the ubiquitin, thus leading to higher levels of Mcl-1 in the cell. Additional information can be found here [[User:Karsten Theis/5B5Q]] | ||
| + | |||
| + | SEE ALSO [[Tumor susceptibility gene 101]] | ||
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
3D structures of ubiqitin
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987 Apr 5;194(3):531-44. PMID:3041007
- ↑ 2.0 2.1 Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ. Three-dimensional structure of ubiquitin at 2.8 A resolution. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3582-5. PMID:2987935
- ↑ Cox MJ, Haas AL, Wilkinson KD. Role of ubiquitin conformations in the specificity of protein degradation: iodinated derivatives with altered conformations and activities. Arch Biochem Biophys. 1986 Nov 1;250(2):400-9. PMID:3022650
- ↑ Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997 Dec;11(14):1245-56. PMID:9409543
- ↑ Hochstrasser, M. 1996. Ubiquitin-dependent protein Degradation. Annu Rev Genet. 30: 405-439
- ↑ Snyder PM, Price MP, McDonald FJ, Adams CM, Volk KA, Zeiher BG, Stokes JB, Welsh MJ. Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na+ channel. Cell. 1995 Dec 15;83(6):969-78. PMID:8521520
Proteopedia Page Contributors and Editors (what is this?)
Jaclyn Gordon, Joel L. Sussman, Michal Harel, David Canner, Andrea Gorrell, Alexander Berchansky, Karsten Theis