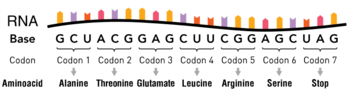Transfer RNA (tRNA)
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 16: | Line 16: | ||
</jmolLink> | </jmolLink> | ||
</jmol> on the other end. | </jmol> on the other end. | ||
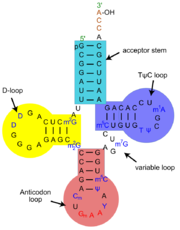
| - | At the acceptor end, amino acid are attached via the <scene name='43/433638/Threeprime/3'>2'-OH or 3'-OH group of the last nucleotide in the acceptor stem</scene>. At the opposite end of the molecule is the anticodon, which pairs with its complementary codon on the messenger RNA. | + | At the acceptor end, amino acid are attached via the <scene name='43/433638/Threeprime/3'>2'-OH or 3'-OH group of the last nucleotide in the acceptor stem</scene>. At the opposite end of the molecule is the <scene name='43/433638/Anticodon_loop/1'>anticodon</scene>, which pairs with its complementary codon on the messenger RNA. |
The two arms of the "L" <scene name='43/433638/Fullview_cartoon/20'> (cartoon)</scene> are formed by the <scene name='43/433638/Stemstacking/3'>stacking of the acceptor and TΨC-stem</scene> on one side, and of the anticodon and D-stem on the other side. <scene name='43/433638/Kissing/4'>Tertiary interactions between the TΨC- and D-loop</scene> form the corner of the L-shape and stabilize the structure. Non-Watson-Crick hydrogen bonding is important in this core (visualize interactively at [http://jmol.x3dna.org/ DSSR Jmol web interface)]). | The two arms of the "L" <scene name='43/433638/Fullview_cartoon/20'> (cartoon)</scene> are formed by the <scene name='43/433638/Stemstacking/3'>stacking of the acceptor and TΨC-stem</scene> on one side, and of the anticodon and D-stem on the other side. <scene name='43/433638/Kissing/4'>Tertiary interactions between the TΨC- and D-loop</scene> form the corner of the L-shape and stabilize the structure. Non-Watson-Crick hydrogen bonding is important in this core (visualize interactively at [http://jmol.x3dna.org/ DSSR Jmol web interface)]). | ||
| Line 36: | Line 36: | ||
See also [[Transfer RNA tour]]. | See also [[Transfer RNA tour]]. | ||
| - | </StructureSection> | ||
==Function== | ==Function== | ||
| Line 49: | Line 48: | ||
'''Aminoacylation.''' Aminoacyl tRNA Synthetases pair amino acids with tRNAs. In this way, they implement the genetic code. These enzymes recognize a single tRNA (e.g. phe-tRNA) and a single amino acid (phenylalanine, in this example) and catalyze formation of an ester bond between the 3’-hydroxyl of the tRNA and the carboxylatic acid of the amino acid. | '''Aminoacylation.''' Aminoacyl tRNA Synthetases pair amino acids with tRNAs. In this way, they implement the genetic code. These enzymes recognize a single tRNA (e.g. phe-tRNA) and a single amino acid (phenylalanine, in this example) and catalyze formation of an ester bond between the 3’-hydroxyl of the tRNA and the carboxylatic acid of the amino acid. | ||
<scene name='43/433638/Cv/4'>Glutaminyl-tRNA synthetase/tRNA complex (1gtr)</scene>. The cognate aminoacid is esterified on its 3'-OH by the cognate aminoacyl-tRNA synthetase. The synthetase recognizes structural features on the tRNA, which allows it to discriminate tRNA that are specific for a given aminoacid, from all other (non-cognate) tRNA. These structural features are called identity determinants. They are often (but not exclusively) located in the anticodon sequence and/or in the so-called discriminator base (position 73), just before the 3' -CCA terminus. | <scene name='43/433638/Cv/4'>Glutaminyl-tRNA synthetase/tRNA complex (1gtr)</scene>. The cognate aminoacid is esterified on its 3'-OH by the cognate aminoacyl-tRNA synthetase. The synthetase recognizes structural features on the tRNA, which allows it to discriminate tRNA that are specific for a given aminoacid, from all other (non-cognate) tRNA. These structural features are called identity determinants. They are often (but not exclusively) located in the anticodon sequence and/or in the so-called discriminator base (position 73), just before the 3' -CCA terminus. | ||
| + | |||
| + | </StructureSection> | ||
__NOTOC__ | __NOTOC__ | ||
| Line 253: | Line 254: | ||
* [[Translation]] | * [[Translation]] | ||
* [[Ribosome]] | * [[Ribosome]] | ||
| + | * [[Pseudouridine]] | ||
* [[2czj|tmRNA]] | * [[2czj|tmRNA]] | ||
| + | * [[Aminoacyl tRNA Synthetase]] | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Current revision
tRNA or transfer RNA plays a key role in translation, the process of synthesizing proteins from amino acids in a sequence specified by information contained in messenger RNA[1][2]. During this process, triplets of nucleotides (codons) of the messenger RNA are translated according to the genetic code into one of the 20 amino acids. tRNAs serve as the dictionary in this translation process. They contain a specific triplet nucleotide sequence, the anticodon, and they get attached to a specific (cognate) amino acid. During protein synthesis by ribosomes, tRNAs deliver the correct amino acids through interactions of their anticodon region with the complementary codons on the messenger RNA. Apart from their distinct anticodon regions, different tRNAs have very similar structures, allowing them to all fit into the tRNA-binding sites on the ribosome.
| |||||||||||
3D Structures of tRNA
Updated on 23-September-2022
See Also
References
- ↑ Biochemistry 5th ed. Berg JM, Tymoczko JL, Stryer L. New York: W H Freeman; 2002. Section 29.1 "Protein Synthesis Requires the Translation of Nucleotide Sequences Into Amino Acid Sequences" retrieved on 10/31/2018 from [1]
- ↑ Molecular Biology of the Cell. 4th ed. Section "From RNA to Protein" retrieved on 10/31/2018 from [2]
- ↑ Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435-40. PMID:4601792
- ↑ Kim SH, Sussman JL, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC, RICH A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4970-4. PMID:4612535
- ↑ Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010 Jun 22;49(24):4934-44. PMID:20459084 doi:10.1021/bi100408z
Reference for the structure
- Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000 Aug;6(8):1091-105. PMID:10943889
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Wayne Decatur, Michal Harel, Frédéric Dardel, Ann Taylor, Joel L. Sussman, Alexander Berchansky
Categories: Trna | Topic Page | Translation | Modification | RNA | Amino acid