Sandbox Reserved 1507
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 12: | Line 12: | ||
---- | ---- | ||
| - | Hsp70 is a family of proteins. They chaperone plenty of cellular proteins and their role is to help the good folding of proteins by preventing aggregation and degradation and help protein transport across membranes. | + | Hsp70 is a family of proteins. They chaperone plenty of cellular proteins and their role is to help the good folding of proteins by preventing aggregation and degradation and help protein transport across membranes. <ref> DOI: 10.1126/science.1068408 </ref> |
Hsp70 is composed of two chains and is 382 nucleotides long. It’s molecular weight is 70 kDa. It can be linked to different ligands such as ATP and ions like potassium or magnesium. <ref>DOI 10.1016/j.cell.2008.05.022</ref> | Hsp70 is composed of two chains and is 382 nucleotides long. It’s molecular weight is 70 kDa. It can be linked to different ligands such as ATP and ions like potassium or magnesium. <ref>DOI 10.1016/j.cell.2008.05.022</ref> | ||
| Line 18: | Line 18: | ||
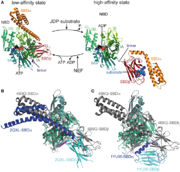
The aim of the NBD is to regulate the activity of PDB for the substrate binding and release. | The aim of the NBD is to regulate the activity of PDB for the substrate binding and release. | ||
| - | Conformational cycle of Hsp70s <ref> doi: 10.3389/fmolb.2015.00058 </ref> | + | |
| - | + | [[Image:Hsp70.jpg | thumb | Conformational cycle of Hsp70s <ref> doi: 10.3389/fmolb.2015.00058 </ref> ]] | |
Hsp70s recognizes a short degenerate sequence motif, present in most polypeptides. | Hsp70s recognizes a short degenerate sequence motif, present in most polypeptides. | ||
| Line 31: | Line 31: | ||
The Hsp110 family is composed of two proteins Sse1p and Sse2p. Sse1p is constitutively expressed while the production of Sse2p is stress dependant. | The Hsp110 family is composed of two proteins Sse1p and Sse2p. Sse1p is constitutively expressed while the production of Sse2p is stress dependant. | ||
| - | Hsp110 proteins are homologous to Hsp70. The general domain organisation of Sse1p ressemble to one of the canonical Hsp70s. They are composed of different domains: a N-terminal actin type Nucleotide-Binding Domain, a ß sandwich domain and a C-terminal 3 helix bundle domain (3HBD). | + | Hsp110 proteins <ref> DOI: 10.1515/hsz-2018-0209 </ref> are homologous to Hsp70. The general domain organisation of Sse1p ressemble to one of the canonical Hsp70s. They are composed of different domains: a N-terminal actin type Nucleotide-Binding Domain, a ß sandwich domain and a C-terminal 3 helix bundle domain (3HBD). |
All the interaction with the NBD of Hsp70 is located along the NBD and 3HBD of the Sse1p. The interaction with Hsp70 only causes minor changes in its conformation. | All the interaction with the NBD of Hsp70 is located along the NBD and 3HBD of the Sse1p. The interaction with Hsp70 only causes minor changes in its conformation. | ||
| Line 53: | Line 53: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | <references> | ||
Current revision
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
3D2F, Crystal structure of a complex of Sse1p and Hsp70
| |||||||||||
References
- ↑ Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002 Mar 8;295(5561):1852-8. doi: 10.1126/science.1068408. PMID:11884745 doi:http://dx.doi.org/10.1126/science.1068408
- ↑ Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008 Jun 13;133(6):1068-79. PMID:18555782 doi:10.1016/j.cell.2008.05.022
- ↑ Mayer MP, Kityk R. Insights into the molecular mechanism of allostery in Hsp70s. Front Mol Biosci. 2015 Oct 20;2:58. doi: 10.3389/fmolb.2015.00058. eCollection, 2015. PMID:26539440 doi:http://dx.doi.org/10.3389/fmolb.2015.00058
- ↑ Yakubu UM, Morano KA. Roles of the nucleotide exchange factor and chaperone Hsp110 in cellular proteostasis and diseases of protein misfolding. Biol Chem. 2018 Sep 25;399(10):1215-1221. doi: 10.1515/hsz-2018-0209. PMID:29908125 doi:http://dx.doi.org/10.1515/hsz-2018-0209
- ↑ Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008 Jun 13;133(6):1068-79. PMID:18555782 doi:10.1016/j.cell.2008.05.022