User:Estelle Blochouse/ Sandbox 1497
From Proteopedia
< User:Estelle Blochouse(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 19: | Line 19: | ||
Multicopper oxidase catalyzes the oxidation of different substrates by reducing O2 into H2O without releasing activated oxygen species (H2O2). | Multicopper oxidase catalyzes the oxidation of different substrates by reducing O2 into H2O without releasing activated oxygen species (H2O2). | ||
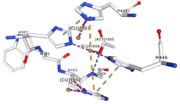
| - | Three different copper centers exist | + | Three different copper centers exist which can be differentiated by spectroscopy: Type 1 or blue (<scene name='pdbligand=CU:COPPER+(II)+ION'>mononoclear copper center</scene>), type 2 or normal and type 3 or coupled binuclear.<ref>Messerschmidt A, Huber R, The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships, Eur. J. Biochem. 187, January 1990</ref><ref>Ouzounis C, Sander C, A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins, FEBS Lett. volume 279, February 1991</ref>. |
| - | Multicopper oxidase contains these 3 types of copper ions. They are all involved in the transfer of electrons from the substrate to the dioxygen. The first type of copper, type 1 | + | Multicopper oxidase contains these 3 types of copper ions. They are all involved in the transfer of electrons from the substrate to the dioxygen. The first type of copper, type 1 (Cu1001) mediates the electron transfer from the substrate to the other coppers. The <scene name='pdbligand=CU:COPPER+(II)+ION'>mononoclear copper center</scene> formed by the three other coppers is the key element for the oxygen reduction. It contains a type 2 copper (Cu1004) and two type 3 copper ions, binuclear ions (Cu1002 and Cu1003). The final electron acceptor, O2, is bound to this last type of copper and is reduced into two molecules of water thanks to the transfer of four electrons allowed by the oxidation of the four copper ions.<ref>Hirofumi Komori, Ryosuke |
Sugiyama, Kunishige Kataoka, | Sugiyama, Kunishige Kataoka, | ||
Kentaro Miyazaki, Yoshiki | Kentaro Miyazaki, Yoshiki | ||
| Line 28: | Line 28: | ||
== Disease == | == Disease == | ||
| - | If the amino acids 500 and 501 are mutated from CH to SR, | + | If the amino acids 500 and 501 are mutated from CH to SR, there is a loss of resistance to copper which is highly harmful for the bacteria. |
Oxygen deprivation also leads to protein degradation. | Oxygen deprivation also leads to protein degradation. | ||
| Line 53: | Line 53: | ||
Even if the trinuclear copper center is deeply buried inside the protein, the channel is dynamic enough to allow the acetate ion to approach. It has been noticed that adding acetate ion is enhancing the enzymatic activity of the protein. <br> | Even if the trinuclear copper center is deeply buried inside the protein, the channel is dynamic enough to allow the acetate ion to approach. It has been noticed that adding acetate ion is enhancing the enzymatic activity of the protein. <br> | ||
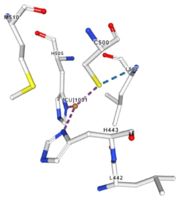
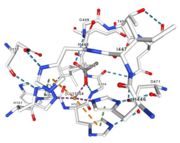
</td></tr><tr><td>[[Image:Cu1001.jpg|thumb|left|'''Figure 2:''' Copper 1001<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1001: The copper ion is bound with H443, H505 and C500 thanks to three metallic interactions. The structure is stabilised by hydrogen bounds with L502, M510.</td></tr> | </td></tr><tr><td>[[Image:Cu1001.jpg|thumb|left|'''Figure 2:''' Copper 1001<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1001: The copper ion is bound with H443, H505 and C500 thanks to three metallic interactions. The structure is stabilised by hydrogen bounds with L502, M510.</td></tr> | ||
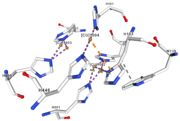
| - | <tr><td>[[Image:Cu1002.jpg|thumb|left|'''Figure 3:''' Copper 1002<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1002: The binuclear copper ion is bound with | + | <tr><td>[[Image:Cu1002.jpg|thumb|left|'''Figure 3:''' Copper 1002<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1002: The binuclear copper ion is bound with three histidines : H501, H103, and H141. Each bound is a double metallic interaction because the two nucleus of the binuclear ion are bound. The structure is stabilised by hydrophobic contact between H141 and W139</td></tr> |
<tr><td>[[Image:Cu1003.jpg|thumb|left|'''Figure 4:''' Copper 1003<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1003: The binuclear copper ion is bound with three with three histidines : H499, H143, and H448. Each bound is a double metallic interaction. The structure is stabilised by Pi interactions with Cu1004 and H101</td></tr> | <tr><td>[[Image:Cu1003.jpg|thumb|left|'''Figure 4:''' Copper 1003<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1003: The binuclear copper ion is bound with three with three histidines : H499, H143, and H448. Each bound is a double metallic interaction. The structure is stabilised by Pi interactions with Cu1004 and H101</td></tr> | ||
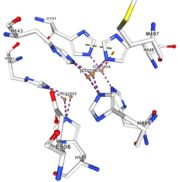
| - | <tr><td>[[Image:Cu1004.jpg|thumb|left|'''Figure 5:''' Copper 1004<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1004: The copper ion is bound by metallic interaction with two histidines : H101 and H446, and with Act1005. The structure is stabilised by Pi interactions between H446 and two other | + | <tr><td>[[Image:Cu1004.jpg|thumb|left|'''Figure 5:''' Copper 1004<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Cu1004: The copper ion is bound by metallic interaction with two histidines : H101 and H446, and with Act1005. The structure is stabilised by Pi interactions between H446 and two other histidines : H103 and H448. Cu1004 bounds also by Pi interactions with H103 and H448. Moreover, their are Pi interactions between H113 and H446</td></tr> |
<tr><td>[[Image:Act10051.jpg|thumb|left|'''Figure 6:''' Acetate ion 1005<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Act1005: The acetate ion is linked by hydrogen bonds to G104 and G449.</td></tr> | <tr><td>[[Image:Act10051.jpg|thumb|left|'''Figure 6:''' Acetate ion 1005<ref><span class='plainlinks'>[https://www.rcsb.org/structure/4e9s RCBS PDB]</span></ref>]]</td><td>Act1005: The acetate ion is linked by hydrogen bonds to G104 and G449.</td></tr> | ||
</table> | </table> | ||
| - | Cu1002, Cu1003, Cu1004 and Act1005 are very closed in the | + | Cu1002, Cu1003, Cu1004 and Act1005 are very closed in the tridimensionnal structure of the protein. The three copper ions form the trinuclear copper center. The bound lengths are nearly equal so the three copper are arranged as a triangle. |
Current revision
Multicopper Oxidase CueO (4e9s)
| |||||||||||
References
- ↑ EMBL-EBI, Family: Cu-oxidase (PF00394), Summary: Multicopper oxidase, http://pfam.xfam.org/family/Cu-oxidase
- ↑ UniProtKB
- ↑ Messerschmidt A, Huber R, The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships, Eur. J. Biochem. 187, January 1990
- ↑ Ouzounis C, Sander C, A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins, FEBS Lett. volume 279, February 1991
- ↑ Hirofumi Komori, Ryosuke Sugiyama, Kunishige Kataoka, Kentaro Miyazaki, Yoshiki Higuchib, and Takeshi Sakurai, New insights into the catalytic active-site structure of multicopper oxidases, Biological Crystallography, 6 December 2013 doi:10.1107/S1399004713033051
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ Kataoka K, Komori H, Ueki Y, Konno Y, Kamitaka Y, Kurose S, Tsujimura S, Higuchi Y, Kano K, Seo D, Sakurai T. Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site. J Mol Biol. 2007 Oct 12;373(1):141-52. Epub 2007 Aug 2. PMID:17804014 doi:10.1016/j.jmb.2007.07.041