We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Frataxin
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 12: | Line 12: | ||
In the box at the right, it is possible to see its <scene name='78/788815/Spacefill_model/1'>general structure</scene> in a space-fill model, in which <font color='violet'><b>violet</b></font>, <font color='orangered'><b>orange</b></font> and <span style="color:aquamarine;background-color:darkgrey;font-weight:bold;">light-green</span> represent, each, a different monomer from the entire molecule. | In the box at the right, it is possible to see its <scene name='78/788815/Spacefill_model/1'>general structure</scene> in a space-fill model, in which <font color='violet'><b>violet</b></font>, <font color='orangered'><b>orange</b></font> and <span style="color:aquamarine;background-color:darkgrey;font-weight:bold;">light-green</span> represent, each, a different monomer from the entire molecule. | ||
However, to cover some important aspects of the structure and function of the molecule, it is particularly useful to represent its <scene name='78/788815/Secondary_structure/1'>secondary structure patterns</scene>. | However, to cover some important aspects of the structure and function of the molecule, it is particularly useful to represent its <scene name='78/788815/Secondary_structure/1'>secondary structure patterns</scene>. | ||
| + | |||
| + | '''CyaY''' is a bacterial frataxin ortholog which participates in iron-sulfur assembly as an iron-dependent inhibitor of cluster formation<ref>PMID:19305405</ref>. The yeast '''frataxin homolog FH1''' plays a role in maturation of iron-sulfur proteins<ref>PMID:12165564</ref>. | ||
---- | ---- | ||
| Line 74: | Line 76: | ||
== Disease == | == Disease == | ||
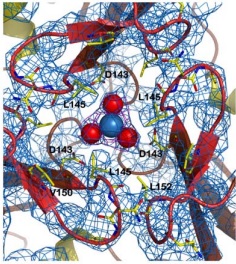
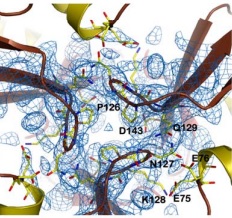
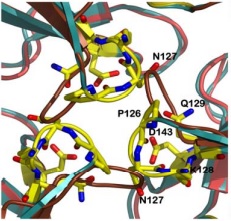
| - | Friedreich ataxia is developed when the frataxin trimer cannot stabilize itself to perform the ferroxidase activity, with is probably dependent on conformational changes that take place when the monomers come together, being in a site around the residues H74, D78, D79, D82 and H83. | + | Friedreich ataxia<ref>PMID:18852343</ref> is developed when the frataxin trimer cannot stabilize itself to perform the ferroxidase activity, with is probably dependent on conformational changes that take place when the monomers come together, being in a site around the residues H74, D78, D79, D82 and H83. |
There are 4 know mutations capable of creating such defects: <scene name='78/786054/Mutation_107_123/1'>G107V</scene>, that breaks the hydrogen bond between G107 and K123, destabilizing the region of 123-130; W131, necessary to stabilize the N-terminal extension; <scene name='78/786054/Mutation_141-125/1'>R141C</scene> breaks an hydrogen bound between R141-P125, which may affect the conformation of the 125-128 loop and D122Y | There are 4 know mutations capable of creating such defects: <scene name='78/786054/Mutation_107_123/1'>G107V</scene>, that breaks the hydrogen bond between G107 and K123, destabilizing the region of 123-130; W131, necessary to stabilize the N-terminal extension; <scene name='78/786054/Mutation_141-125/1'>R141C</scene> breaks an hydrogen bound between R141-P125, which may affect the conformation of the 125-128 loop and D122Y | ||
| - | </StructureSection> | ||
== 3D Structures of frataxin == | == 3D Structures of frataxin == | ||
| - | + | [[Frataxin 3D Structures]] | |
| - | + | ||
| - | + | </StructureSection> | |
| - | **[[1ekg]], [[3s4m]] – hFXN C-terminal - human<br /> | ||
| - | **[[1ly7]] – hFXN C-terminal - NMR<br /> | ||
| - | **[[3s5d]], [[3s5e]], [[3s5f]], [[3t3j]], [[3t3k]], [[3t3l]], [[3t3t]], [[3t3x]] – hFXN C-terminal (mutant)<br /> | ||
| - | **[[5kz5]] – hFXN C-terminal + iron sulfur assembly protein + cysteine desulfarase – Cryo EM<br /> | ||
| - | |||
| - | *FXN homolog; yeast residues 53-174 | ||
| - | |||
| - | **3co6, 3coa, [[3co7]], [[2ga5]] – yFH1 – yeast – NMR<br /> | ||
| - | **[[2fql]], [[3oeq]] – yFH1 (mutant)<br /> | ||
| - | **[[3oer]] – yFH1 (mutant) + Co<br /> | ||
| - | **[[4ec2]] – yFH1 (mutant) + Fe<br /> | ||
| - | **[[5tre]], [[5t0v]] – yFH1 + iron sulfur assembly protein – Cryo EM<br /> | ||
| - | |||
| - | *FXN ortholog CyaA | ||
| - | |||
| - | **[[1ew4]] – EcCyaY – Escherichia coli <br /> | ||
| - | **[[1soy]] – EcCyaY – NMR<br /> | ||
| - | **[[2eff]] – EcCyaY + Co <br /> | ||
| - | **[[2p1x]] – EcCyaY + Eu <br /> | ||
| - | **[[4jpd]] – CyaY – Burkholderia cenocepacia<br /> | ||
| - | **[[4hs5]] – PiCyaY – Psychromonas ingrahamii<br /> | ||
| - | **[[4lk8]] – PiCyaY + Co <br /> | ||
| - | **[[4lp1]] – PiCyaY + Eu<br /> | ||
| - | }} | ||
== References == | == References == | ||
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006. | KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006. | ||
| + | <references/> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006.
- ↑ Adinolfi S, Iannuzzi C, Prischi F, Pastore C, Iametti S, Martin SR, Bonomi F, Pastore A. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat Struct Mol Biol. 2009 Apr;16(4):390-6. doi: 10.1038/nsmb.1579. Epub 2009 Mar , 22. PMID:19305405 doi:http://dx.doi.org/10.1038/nsmb.1579
- ↑ Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet. 2002 Aug 15;11(17):2025-36. PMID:12165564
- ↑ Pandolfo M. Friedreich ataxia. Arch Neurol. 2008 Oct;65(10):1296-303. doi: 10.1001/archneur.65.10.1296. PMID:18852343 doi:http://dx.doi.org/10.1001/archneur.65.10.1296
Proteopedia Page Contributors and Editors (what is this?)
João Victor Paccini Coutinho, Michal Harel, Rebeca B. Candia