We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Frataxin
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 12: | Line 12: | ||
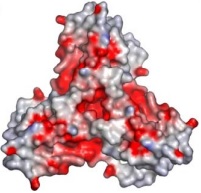
In the box at the right, it is possible to see its <scene name='78/788815/Spacefill_model/1'>general structure</scene> in a space-fill model, in which <font color='violet'><b>violet</b></font>, <font color='orangered'><b>orange</b></font> and <span style="color:aquamarine;background-color:darkgrey;font-weight:bold;">light-green</span> represent, each, a different monomer from the entire molecule. | In the box at the right, it is possible to see its <scene name='78/788815/Spacefill_model/1'>general structure</scene> in a space-fill model, in which <font color='violet'><b>violet</b></font>, <font color='orangered'><b>orange</b></font> and <span style="color:aquamarine;background-color:darkgrey;font-weight:bold;">light-green</span> represent, each, a different monomer from the entire molecule. | ||
However, to cover some important aspects of the structure and function of the molecule, it is particularly useful to represent its <scene name='78/788815/Secondary_structure/1'>secondary structure patterns</scene>. | However, to cover some important aspects of the structure and function of the molecule, it is particularly useful to represent its <scene name='78/788815/Secondary_structure/1'>secondary structure patterns</scene>. | ||
| + | |||
| + | '''CyaY''' is a bacterial frataxin ortholog which participates in iron-sulfur assembly as an iron-dependent inhibitor of cluster formation<ref>PMID:19305405</ref>. The yeast '''frataxin homolog FH1''' plays a role in maturation of iron-sulfur proteins<ref>PMID:12165564</ref>. | ||
---- | ---- | ||
| Line 78: | Line 80: | ||
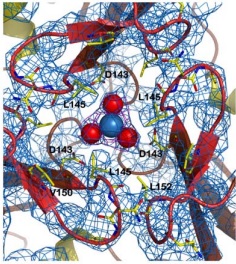
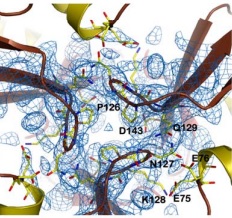
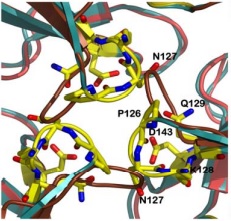
There are 4 know mutations capable of creating such defects: <scene name='78/786054/Mutation_107_123/1'>G107V</scene>, that breaks the hydrogen bond between G107 and K123, destabilizing the region of 123-130; W131, necessary to stabilize the N-terminal extension; <scene name='78/786054/Mutation_141-125/1'>R141C</scene> breaks an hydrogen bound between R141-P125, which may affect the conformation of the 125-128 loop and D122Y | There are 4 know mutations capable of creating such defects: <scene name='78/786054/Mutation_107_123/1'>G107V</scene>, that breaks the hydrogen bond between G107 and K123, destabilizing the region of 123-130; W131, necessary to stabilize the N-terminal extension; <scene name='78/786054/Mutation_141-125/1'>R141C</scene> breaks an hydrogen bound between R141-P125, which may affect the conformation of the 125-128 loop and D122Y | ||
| - | </StructureSection> | ||
== 3D Structures of frataxin == | == 3D Structures of frataxin == | ||
| - | + | [[Frataxin 3D Structures]] | |
| - | + | ||
| - | + | </StructureSection> | |
| - | **[[1ekg]], [[3s4m]] – hFXN C-terminal - human<br /> | ||
| - | **[[1ly7]] – hFXN C-terminal - NMR<br /> | ||
| - | **[[3s5d]], [[3s5e]], [[3s5f]], [[3t3j]], [[3t3k]], [[3t3l]], [[3t3t]], [[3t3x]] – hFXN C-terminal (mutant)<br /> | ||
| - | **[[5kz5]] – hFXN C-terminal + iron sulfur assembly protein + cysteine desulfarase – Cryo EM<br /> | ||
| - | |||
| - | *FXN homolog; yeast residues 53-174 | ||
| - | |||
| - | **3co6, 3coa, [[3co7]], [[2ga5]] – yFH1 – yeast – NMR<br /> | ||
| - | **[[2fql]], [[3oeq]] – yFH1 (mutant)<br /> | ||
| - | **[[3oer]] – yFH1 (mutant) + Co<br /> | ||
| - | **[[4ec2]] – yFH1 (mutant) + Fe<br /> | ||
| - | **[[5tre]], [[5t0v]] – yFH1 + iron sulfur assembly protein – Cryo EM<br /> | ||
| - | |||
| - | *FXN ortholog CyaA | ||
| - | |||
| - | **[[1ew4]] – EcCyaY – Escherichia coli <br /> | ||
| - | **[[1soy]] – EcCyaY – NMR<br /> | ||
| - | **[[2eff]] – EcCyaY + Co <br /> | ||
| - | **[[2p1x]] – EcCyaY + Eu <br /> | ||
| - | **[[4jpd]] – CyaY – Burkholderia cenocepacia<br /> | ||
| - | **[[4hs5]] – PiCyaY – Psychromonas ingrahamii<br /> | ||
| - | **[[4lk8]] – PiCyaY + Co <br /> | ||
| - | **[[4lp1]] – PiCyaY + Eu<br /> | ||
| - | }} | ||
== References == | == References == | ||
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006. | KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006. | ||
| + | <references/> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006.
- ↑ Adinolfi S, Iannuzzi C, Prischi F, Pastore C, Iametti S, Martin SR, Bonomi F, Pastore A. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat Struct Mol Biol. 2009 Apr;16(4):390-6. doi: 10.1038/nsmb.1579. Epub 2009 Mar , 22. PMID:19305405 doi:http://dx.doi.org/10.1038/nsmb.1579
- ↑ Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet. 2002 Aug 15;11(17):2025-36. PMID:12165564
- ↑ Pandolfo M. Friedreich ataxia. Arch Neurol. 2008 Oct;65(10):1296-303. doi: 10.1001/archneur.65.10.1296. PMID:18852343 doi:http://dx.doi.org/10.1001/archneur.65.10.1296
Proteopedia Page Contributors and Editors (what is this?)
João Victor Paccini Coutinho, Michal Harel, Rebeca B. Candia