2-Oxoglutarate Dehydrogenase
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
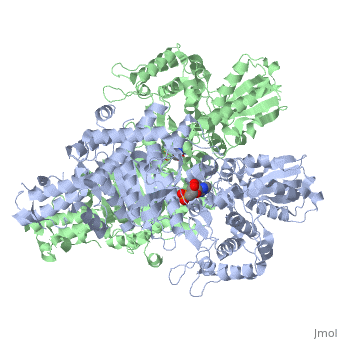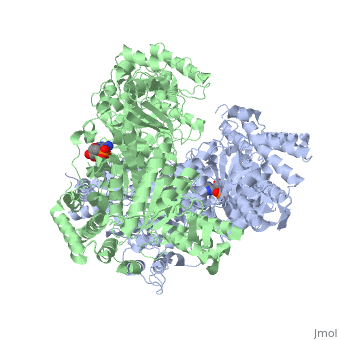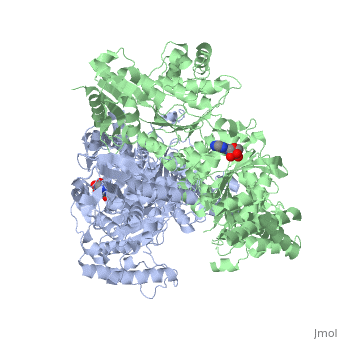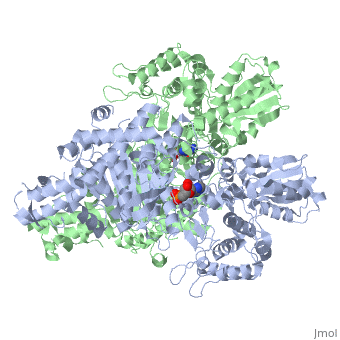
<StructureSection load='2jgd' size='400' side='right' scene='' caption='Dimer of E. coli 2-oxogluterate dehydrogenase E1 component complex with AMP [[2jgd]]'> | <StructureSection load='2jgd' size='400' side='right' scene='' caption='Dimer of E. coli 2-oxogluterate dehydrogenase E1 component complex with AMP [[2jgd]]'> | ||
| + | __TOC__ | ||
| + | ==Introduction== | ||
| + | '''2-Oxoglutarate dehydrogenase''' (OGDH) is an enzyme that plays a crucial role in cellular metabolism. It is part of the tricarboxylic acid (TCA) cycle, also known as the citric acid cycle or Krebs cycle, which is a central metabolic pathway in most organisms. | ||
| + | |||
| + | The OGDH enzyme catalyzes the conversion of 2-oxoglutarate (also known as alpha-ketoglutarate) to succinyl-CoA. This reaction is an important step in the TCA cycle, where 2-oxoglutarate is oxidized, and its energy is harvested in the form of reduced coenzymes, such as NADH and FADH2. | ||
| + | |||
| + | The reaction catalyzed by OGDH involves the decarboxylation of 2-oxoglutarate, resulting in the release of carbon dioxide and the formation of a thioester bond with coenzyme A (CoA). The enzyme complex responsible for this reaction is called the 2-oxoglutarate dehydrogenase complex (OGDHC). OGDHC is similar in structure and function to other multienzyme complexes, such as pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase. | ||
| + | |||
| + | The OGDHC consists of three main components: E1 (2-oxoglutarate dehydrogenase), E2 (dihydrolipoyl succinyltransferase), and E3 (dihydrolipoamide dehydrogenase). Each component plays a specific role in the overall reaction, with E1 catalyzing the decarboxylation of 2-oxoglutarate, E2 transferring the succinyl group to CoA, and E3 regenerating the oxidized form of lipoamide. | ||
| + | The activity of OGDH is regulated through several mechanisms, including allosteric regulation and reversible phosphorylation. Various metabolites, such as NADH, ATP, and succinyl-CoA, can modulate the enzyme's activity, allowing for fine-tuning of the TCA cycle in response to the cell's metabolic needs. | ||
| + | |||
| + | The OGDH enzyme and the TCA cycle are essential for cellular energy production and the generation of metabolic intermediates for biosynthesis. Dysregulation or dysfunction of OGDH can contribute to metabolic disorders and diseases, highlighting the importance of this enzyme in maintaining cellular homeostasis. | ||
| + | |||
| + | ==Key Features== | ||
'''2-oxoglutarate dehydrogenase''' is a complex of three components: E1, E2 and E3. The enzyme '''2-Oxoglutarate Dehydrogenase E1o''' (OGDH) is a subunit the 2-Oxoglutarate Dehydrogenase ([[EC Number]] [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.2.4.2 1.2.4.2]) multi enzyme complex. This subunit is a homo-dimer and one of three enzymes that make up the multi-enzyme complex of 2-Oxoglutarate Dehydrogenase. Each of the <scene name='Lucas_Evans_Sandbox/Monomer/2'>monomers</scene> that make up the homo-dimer have an <scene name='Lucas_Evans_Sandbox/Amp1/1'>adenosine monophosphate</scene> cofactor that facilitates the catalysis. E1o is not categorized in the Structural Classification of Proteins (SCOP); however, the <scene name='Lucas_Evans_Sandbox/Secondary_structure/2'>secondary structure</scene> of one of the dimers shows that this enzyme has large sections of alpha-helices followed by a large sections of parallel beta-pleated sheets these alpha and beta subunits are fused as a single polypeptide <ref name="one">PMID:17367808 </ref>. E2 component is '''dihydrolipoyl succinyltransferase''' or '''dihydrolipoamide succinyltransferase''' (DLST) with lipoic acid as coenzyme and E3 is [[Dihydrolipoamide dehydrogenase]] (DLD) with FAD as coenzyme. The 2-oxoglutaqrate dehydrogenase complex participates in then citric acid cycle, lysine degradation and tryptophan metabolism. | '''2-oxoglutarate dehydrogenase''' is a complex of three components: E1, E2 and E3. The enzyme '''2-Oxoglutarate Dehydrogenase E1o''' (OGDH) is a subunit the 2-Oxoglutarate Dehydrogenase ([[EC Number]] [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.2.4.2 1.2.4.2]) multi enzyme complex. This subunit is a homo-dimer and one of three enzymes that make up the multi-enzyme complex of 2-Oxoglutarate Dehydrogenase. Each of the <scene name='Lucas_Evans_Sandbox/Monomer/2'>monomers</scene> that make up the homo-dimer have an <scene name='Lucas_Evans_Sandbox/Amp1/1'>adenosine monophosphate</scene> cofactor that facilitates the catalysis. E1o is not categorized in the Structural Classification of Proteins (SCOP); however, the <scene name='Lucas_Evans_Sandbox/Secondary_structure/2'>secondary structure</scene> of one of the dimers shows that this enzyme has large sections of alpha-helices followed by a large sections of parallel beta-pleated sheets these alpha and beta subunits are fused as a single polypeptide <ref name="one">PMID:17367808 </ref>. E2 component is '''dihydrolipoyl succinyltransferase''' or '''dihydrolipoamide succinyltransferase''' (DLST) with lipoic acid as coenzyme and E3 is [[Dihydrolipoamide dehydrogenase]] (DLD) with FAD as coenzyme. The 2-oxoglutaqrate dehydrogenase complex participates in then citric acid cycle, lysine degradation and tryptophan metabolism. | ||
See also | See also | ||
| Line 9: | Line 23: | ||
*[[Citric Acid Cycle]] | *[[Citric Acid Cycle]] | ||
*[[Krebs cycle step 4]]. | *[[Krebs cycle step 4]]. | ||
| - | + | ||
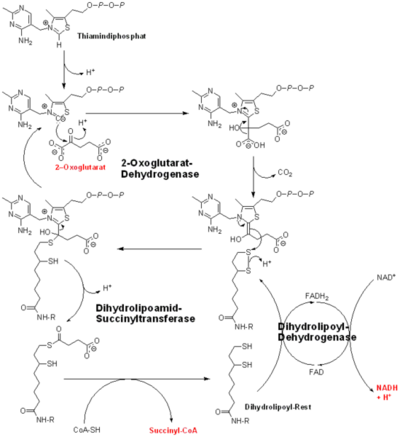
==Catalysis== | ==Catalysis== | ||
[[Image:oxoglutarate dehydrogenase.png|thumb|left|400px|The complete reaction of the Oxoglutarate Dehydrogenase Multi-Enzyme Complex]] | [[Image:oxoglutarate dehydrogenase.png|thumb|left|400px|The complete reaction of the Oxoglutarate Dehydrogenase Multi-Enzyme Complex]] | ||
| Line 27: | Line 41: | ||
</StructureSection> | </StructureSection> | ||
| - | == 3D structures of 2-Oxoglutarate dehydrogenase== | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *E1 component | ||
| - | |||
| - | **[[2jgd]] - Ec2OGDH E1 - ''Escherichia coli'' <br /> | ||
| - | |||
| - | *E2 component | ||
| - | |||
| - | **[[1bal]], [[1bbl]], [[2btg]], [[2bth]], [[2cyu]] - EcDLST E3-binding domain - NMR <br /> | ||
| - | **[[1ghj]], [[1ghk]] - AvDLST lipoyl-binding domain - NMR - ''Azotobacter vinelandii'' <br /> | ||
| - | **[[1pmr]] - EcDLST lipoyl-binding domain - NMR<br /> | ||
| - | **[[1e2o]], [[1c4t]], [[1scz]] - EcDLST catalytic domain<br /> | ||
| - | **[[2eq7]] – Tt2OGDH E3+E2 – ''Thermus thermophilus''<br /> | ||
| - | |||
| - | *E3 component | ||
| - | |||
| - | **[[Dihydrolipoamide dehydrogenase]] | ||
| - | |||
| - | }} | ||
==References== | ==References== | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Frank RA, Price AJ, Northrop FD, Perham RN, Luisi BF. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J Mol Biol. 2007 May 4;368(3):639-51. Epub 2007 Feb 7. PMID:17367808 doi:10.1016/j.jmb.2007.01.080
- ↑ 2.0 2.1 McMinn CL, Ottaway JH. Studies on the mechanism and kinetics of the 2-oxoglutarate dehydrogenase system from pig heart. Biochem J. 1977 Mar 1;161(3):569-81. PMID:192200
- ↑ Leung PS, Rossaro L, Davis PA, Park O, Tanaka A, Kikuchi K, Miyakawa H, Norman GL, Lee W, Gershwin ME. Antimitochondrial antibodies in acute liver failure: implications for primary biliary cirrhosis. Hepatology. 2007 Nov;46(5):1436-42. PMID:17657817 doi:10.1002/hep.21828
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Lucas Evans, Alexander Berchansky, Joel L. Sussman