We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Mark Macbeth/Sandbox5
From Proteopedia
< User:Mark Macbeth(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='1B7F' size='350' frame='true' side='right' caption='Sex lethal (PDB: 1B7F)' scene='78/782600/Opening_image/2'> | <StructureSection load='1B7F' size='350' frame='true' side='right' caption='Sex lethal (PDB: 1B7F)' scene='78/782600/Opening_image/2'> | ||
== Introduction == | == Introduction == | ||
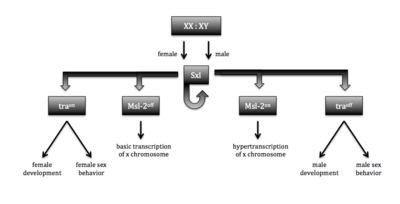
| - | [[Image:Sxl function 2018-03-27 at 2.41.00 PM.png| | + | [[Image:Sxl function 2018-03-27 at 2.41.00 PM.png|400 px|right|thumb|Figure 1. Sxl sex determination and dosage compensation pathways. Functionality of Sxl is determined by the number of X chromosomes. The functional protein (XX) causes a cascade that leads to female structures and behaviors, while the nonfunctional protein results in the default male structures and behaviors.]]Sex lethal (Sxl) is an [https://en.wikipedia.org/wiki/RNA-binding_protein RNA-binding protein] that plays a vital role in sex determination and dosage compensation in ''[https://en.wikipedia.org/wiki/Drosophila_melanogaster Drosophila melanogaster]'', the common fruit fly <ref name="Black">doi: 10.1146/annurev.biochem.72.121801.161720</ref>. Sxl binds specifically to the continuous single-stranded RNA sequence 5’-UGUUUUUUU <ref name="Handa">doi:10.1038/19242 |
</ref>. Functional copies of Sxl are expressed only in female fruit flies, where they induce sex-specific splicing patterns in the transcript of the ''[https://en.wikipedia.org/wiki/Transformer_(gene) Transformer]'' (Tra) gene that lead to its function. Tra initiates a cascade pathway that results in the development of female structures and behaviors (Figure 1). Sxl binds to its recognition element in the Tra pre-mRNA transcript, thereby blocking association of the splicing factor U2AF at the nearby splice site. Without the association of this essential splicing factor, the 3’ splice site shifts downstream, causing the removal of a premature stop codon and preventing truncation and inactivation of the Tra protein <ref name="Black" />. The pre-mRNA transcript of ''[https://www.wikigenes.org/e/gene/e/33565.html Male-specific lethal 2]'' (Msl-2) is the downstream target through which Sxl regulates dosage compensation. Active Sxl protein (in females) binds to two recognition elements on Msl-2 causing the retention of the first intron in the 5’UTR of Msl-2. Sxl protein bound at this intron then blocks translation, preventing expression of Msl-2 in females <ref name="Black" />. With no Msl-2 protein present, the Male-specific lethal complex cannot form and carry out its function of upregulating expression of genes on the X chromosome <ref name="dosage">Georgiev P, Chlamydas S, Akhtar A. Drosophila dosage compensation. Fly 2011;5(2):147-154. https://doi.org/10.4161/fly.5.2.14934</ref> (Figure 1). | </ref>. Functional copies of Sxl are expressed only in female fruit flies, where they induce sex-specific splicing patterns in the transcript of the ''[https://en.wikipedia.org/wiki/Transformer_(gene) Transformer]'' (Tra) gene that lead to its function. Tra initiates a cascade pathway that results in the development of female structures and behaviors (Figure 1). Sxl binds to its recognition element in the Tra pre-mRNA transcript, thereby blocking association of the splicing factor U2AF at the nearby splice site. Without the association of this essential splicing factor, the 3’ splice site shifts downstream, causing the removal of a premature stop codon and preventing truncation and inactivation of the Tra protein <ref name="Black" />. The pre-mRNA transcript of ''[https://www.wikigenes.org/e/gene/e/33565.html Male-specific lethal 2]'' (Msl-2) is the downstream target through which Sxl regulates dosage compensation. Active Sxl protein (in females) binds to two recognition elements on Msl-2 causing the retention of the first intron in the 5’UTR of Msl-2. Sxl protein bound at this intron then blocks translation, preventing expression of Msl-2 in females <ref name="Black" />. With no Msl-2 protein present, the Male-specific lethal complex cannot form and carry out its function of upregulating expression of genes on the X chromosome <ref name="dosage">Georgiev P, Chlamydas S, Akhtar A. Drosophila dosage compensation. Fly 2011;5(2):147-154. https://doi.org/10.4161/fly.5.2.14934</ref> (Figure 1). | ||
| Line 14: | Line 14: | ||
===RNA binding=== | ===RNA binding=== | ||
| - | 1. Hydrogen bonding with RNA bases: | + | ====1. Hydrogen bonding with RNA bases:==== |
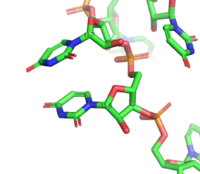
There are numerous hydrogen bonds between the residues and the nucleotide bases. RRM2 creates three hydrogen bonds with U3 and G4 with residues <scene name='78/782600/H-bond_u3_bases/3'>Asn217 and Arg252</scene> and residue <scene name='78/782600/Ala289_g4/1'>Ala289</scene>. It also forms two hydrogen bonds with U5 with residues <scene name='78/782600/Rrm2_res_hbond_w_bases/5'>Aln239 and Asn212</scene>. RRM1 creates five hydrogen bonds with U6, U7, and U8 with residues <scene name='78/782600/Rrm1_res_h-bond_w_bases/6'>Arg195, Gln134, and Ser165</scene>. It also creates seven hydrogens bonds to U9, U10, and U11 with residues <scene name='78/782600/U9_u10/1'>Gly204, Arg202, Lys197</scene> and residues <scene name='78/782600/U11/1'>Arg155 and Asn126</scene><ref name="Handa" />. | There are numerous hydrogen bonds between the residues and the nucleotide bases. RRM2 creates three hydrogen bonds with U3 and G4 with residues <scene name='78/782600/H-bond_u3_bases/3'>Asn217 and Arg252</scene> and residue <scene name='78/782600/Ala289_g4/1'>Ala289</scene>. It also forms two hydrogen bonds with U5 with residues <scene name='78/782600/Rrm2_res_hbond_w_bases/5'>Aln239 and Asn212</scene>. RRM1 creates five hydrogen bonds with U6, U7, and U8 with residues <scene name='78/782600/Rrm1_res_h-bond_w_bases/6'>Arg195, Gln134, and Ser165</scene>. It also creates seven hydrogens bonds to U9, U10, and U11 with residues <scene name='78/782600/U9_u10/1'>Gly204, Arg202, Lys197</scene> and residues <scene name='78/782600/U11/1'>Arg155 and Asn126</scene><ref name="Handa" />. | ||
| - | 2. Hydrogen bonding with the RNA backbone: | + | ====2. Hydrogen bonding with the RNA backbone:==== |
There are an unusual abundance of interactions between the protein and the RNA backbone of the ligand. RRM2 residues <scene name='78/782600/U9_u6_u3_backbone_h_bonds/1'>Tyr214, Asn241, Arg252, Arg258</scene> hydrogen bond to the backbone of U3, U6 and U9. RRM1 residues <scene name='78/782600/Rrm1_h_bond_to_u9_u11_backbone/1'>Asn130 and Arg155</scene><ref name="Handa" /> hydrogen bond to the backbone of U9 and U11. | There are an unusual abundance of interactions between the protein and the RNA backbone of the ligand. RRM2 residues <scene name='78/782600/U9_u6_u3_backbone_h_bonds/1'>Tyr214, Asn241, Arg252, Arg258</scene> hydrogen bond to the backbone of U3, U6 and U9. RRM1 residues <scene name='78/782600/Rrm1_h_bond_to_u9_u11_backbone/1'>Asn130 and Arg155</scene><ref name="Handa" /> hydrogen bond to the backbone of U9 and U11. | ||
| - | 3. Intermolecular stacking: | + | ====3. Intermolecular stacking:==== |
[https://en.wikipedia.org/wiki/Stacking_(chemistry) Intermolecular stacking] between the aromatic side chains and the nucleotide bases also contributes to RNA binding. In RRM2, U3-G4-U5 stack with <scene name='78/782600/Rrm2_residues_stacking/4'>V254, Y214, and F256</scene> respectively. In RRM1, residues <scene name='78/782600/Rrm1_intermolecular_stacking/4'>Y131, R195, N130, F170</scene> are involved in favorable intermolecular stacking of aromatic rings with nucleotides U6-U11<ref name="Handa" />. | [https://en.wikipedia.org/wiki/Stacking_(chemistry) Intermolecular stacking] between the aromatic side chains and the nucleotide bases also contributes to RNA binding. In RRM2, U3-G4-U5 stack with <scene name='78/782600/Rrm2_residues_stacking/4'>V254, Y214, and F256</scene> respectively. In RRM1, residues <scene name='78/782600/Rrm1_intermolecular_stacking/4'>Y131, R195, N130, F170</scene> are involved in favorable intermolecular stacking of aromatic rings with nucleotides U6-U11<ref name="Handa" />. | ||
| Line 38: | Line 38: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| + | ==Student Contributors== | ||
| + | *Jeremy Caylor | ||
| + | *Lauren Ciulla | ||
| + | *Becca Lewis | ||
Current revision
Sex lethal: RNA-binding protein
| |||||||||||
Student Contributors
- Jeremy Caylor
- Lauren Ciulla
- Becca Lewis