Aspartate Aminotransferase
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
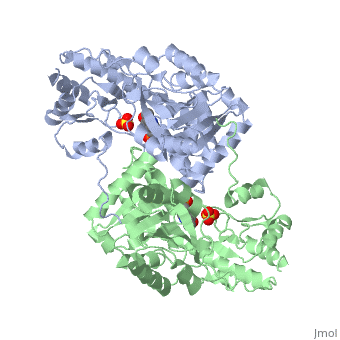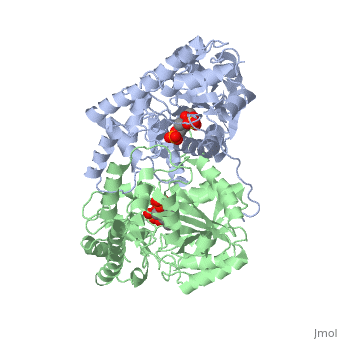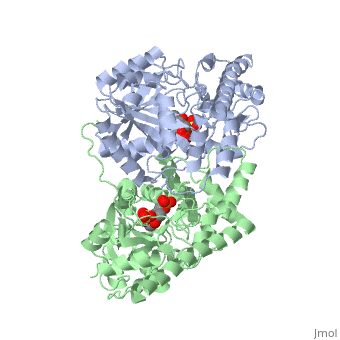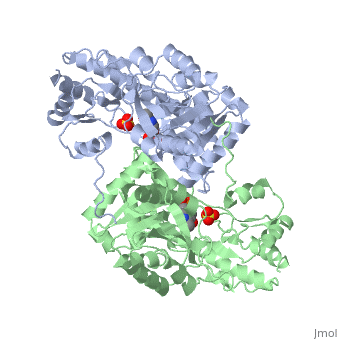
<StructureSection load='1asn' size='400' side='right' scene='' caption='E. coli aspartate aminotransferase complex with PLP and sulfate (PDB code [[1asn]])'> | <StructureSection load='1asn' size='400' side='right' scene='' caption='E. coli aspartate aminotransferase complex with PLP and sulfate (PDB code [[1asn]])'> | ||
| - | + | '''Aspartate Aminotransferase''' | |
by Luke Spooner | by Luke Spooner | ||
| - | '''Aspartate Aminotransferase''' (AAT), also known as '''Glutamic aspartic transaminase''', '''glutamic oxaloacetic transaminase''', '''prephenate aminotransferase''' and '''transaminase A''' is an enzyme that is a member of the class-I pyridoxal-phosphate-dependent aminotransferase family <ref name ="AST family and name">PMID:20977429</ref>. It is coded by the gene GOT1<ref name ="AST gene">PMID:4193185</ref>. It is a homodimer that is 413 amino acids long and serves a critical role in amino acid and carbohydrate metabolism, ureogenesis, and the transfer of reducing equivalents into the mitochondria and chloroplast<ref name ="AST ROLES AND STRUCTURE">PMID:10708649</ref>. Within prokaryote cells it is exclusively found in the cytosol, but in eukaryotic cells there are cytosol, mitochondrial, and chloroplast isozymes<ref name ="AST family and name"/><ref name ="AST Structure"/>. | + | __TOC__ |
| + | |||
| + | ==Function== | ||
| + | |||
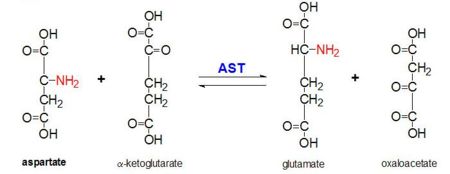
| + | '''Aspartate Aminotransferase''' (AAT), also known as '''Glutamic aspartic transaminase''', '''glutamic oxaloacetic transaminase''', '''prephenate aminotransferase''' and '''transaminase A''' is an enzyme that is a member of the class-I pyridoxal-phosphate-dependent aminotransferase family <ref name ="AST family and name">PMID:20977429</ref>. It is coded by the gene GOT1<ref name ="AST gene">PMID:4193185</ref>. It is a homodimer that is 413 amino acids long and serves a critical role in amino acid and carbohydrate metabolism, ureogenesis, and the transfer of reducing equivalents into the mitochondria and chloroplast<ref name ="AST ROLES AND STRUCTURE">PMID:10708649</ref>. Within prokaryote cells it is exclusively found in the cytosol, but in eukaryotic cells there are cytosol, mitochondrial, and chloroplast isozymes<ref name ="AST family and name"/><ref name ="AST Structure"/>. | ||
| + | *'''Bifunctional aspartate aminotransferase''' (BAAT) is found in plants. It displays aspartate and prephanate aminotransferase activity<ref >PMID:24902885</ref>. | ||
In the human body it is produced in the brain, skeletal muscles, liver, pancreas, red blood cells, and kidneys <ref name ="AST ORGANS">PMID:2569674</ref><ref name ="Liver damage"/>. The wide range of tissues in which it is made, separates it from the similar enzyme alanine transaminase (ALT) which is found primarily in the liver<ref name ="Liver damage">PMID:10831269</ref>. The level of AAT in the body can be used as a marker for tissue disease or damage<ref name ="Liver damage"/>. As well, AAT and ALT levels can be compared to pinpoint whether tissue damage is primarily found within the liver<ref name ="Liver damage">PMID:12546613</ref>. | In the human body it is produced in the brain, skeletal muscles, liver, pancreas, red blood cells, and kidneys <ref name ="AST ORGANS">PMID:2569674</ref><ref name ="Liver damage"/>. The wide range of tissues in which it is made, separates it from the similar enzyme alanine transaminase (ALT) which is found primarily in the liver<ref name ="Liver damage">PMID:10831269</ref>. The level of AAT in the body can be used as a marker for tissue disease or damage<ref name ="Liver damage"/>. As well, AAT and ALT levels can be compared to pinpoint whether tissue damage is primarily found within the liver<ref name ="Liver damage">PMID:12546613</ref>. | ||
| Line 10: | Line 15: | ||
=='''Structure'''== | =='''Structure'''== | ||
<scene name='49/490061/Cv/1'>AST is a homodimer</scene> that contains <scene name='49/490061/Cv/3'>16 α-helices and a β-sheet formed from 7 parallel and antiparallel strands</scene> <ref name ="AST Structure"/> ({{Template:ColorKey_Helix}}, | <scene name='49/490061/Cv/1'>AST is a homodimer</scene> that contains <scene name='49/490061/Cv/3'>16 α-helices and a β-sheet formed from 7 parallel and antiparallel strands</scene> <ref name ="AST Structure"/> ({{Template:ColorKey_Helix}}, | ||
| - | {{Template:ColorKey_Strand}}, {{Template:ColorKey_Loop}}, {{Template:ColorKey_Turn}}). Asymmetric unit of Aspartate aminotransferase, with highlighted small and large domain and PLP cofactor ([[1b4x]]). Each subunit contains an equivalent active site<ref name ="AST Structure">PMID:2121725</ref>. The subunits connect at two sites: between their large domains and between the N-terminal residues and the large domain on the other subunit | + | {{Template:ColorKey_Strand}}, {{Template:ColorKey_Loop}}, {{Template:ColorKey_Turn}}). Asymmetric unit of Aspartate aminotransferase, with highlighted small and large domain and PLP cofactor ([[1b4x]]). Each subunit contains an equivalent active site<ref name ="AST Structure">PMID:2121725</ref>. The subunits connect at two sites: between their large domains and between the N-terminal residues and the large domain on the other subunit. This structure of AST varies minutely among organisms ranging from ''E. coli'' to humans<ref name ="AST Structure"/><ref name ="AST ROLES AND STRUCTURE"/>. As well, the structure of the active site is highly conserved with a sequence homology of 25%<ref name ="AST Structure"/>. |
Each subunit of the homodimer is further divided into a small and large domain<ref name ="AST Structure"/>. The <scene name='Sandbox_Reserved_346/Small_subunit_2/1'>small domain</scene> is comprised of the amino acids from the N-terminus to Pro 48 residue and from Met 326 to the C-terminus<ref name ="AST Structure"/>. The remaining amino acids make up the <scene name='Sandbox_Reserved_346/Large_subunit/1'>large domain</scene>, and the <scene name='Sandbox_Reserved_346/Whole_subunit_2/1'>two domains</scene> are connected by a long α-helix consisting of 32 amino acids<ref name ="AST Structure"/>. | Each subunit of the homodimer is further divided into a small and large domain<ref name ="AST Structure"/>. The <scene name='Sandbox_Reserved_346/Small_subunit_2/1'>small domain</scene> is comprised of the amino acids from the N-terminus to Pro 48 residue and from Met 326 to the C-terminus<ref name ="AST Structure"/>. The remaining amino acids make up the <scene name='Sandbox_Reserved_346/Large_subunit/1'>large domain</scene>, and the <scene name='Sandbox_Reserved_346/Whole_subunit_2/1'>two domains</scene> are connected by a long α-helix consisting of 32 amino acids<ref name ="AST Structure"/>. | ||
| Line 32: | Line 37: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of aspartate aminotransferase== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Aspartate aminotransferase | ||
| - | |||
| - | **[[1aat]] – cAAT – chicken<br /> | ||
| - | **[[3wzf]], [[5ax8]] – hAAT – human<br /> | ||
| - | |||
| - | *Aspartate aminotransferase binary complex with pyridoxamine phosphate | ||
| - | |||
| - | **[[2aat]], [[1aia]], [[1aib]], [[1aic]] – EcAAT (mutant) + pyridoxamine phosphate– ''Escherichia coli''<br /> | ||
| - | **[[4dbc]], [[5t4l]] – EcAAT<br /> | ||
| - | **[[9aat]] - cAAT + pyridoxamine phosphate<br /> | ||
| - | **[[1amq]], [[1amr]], [[1ams]] - EcAAT + pyridoxamine phosphate<br /> | ||
| - | **[[3pd6]] - mAAT + pyridoxamine phosphate - mouse<br /> | ||
| - | **[[2z9u]] – MlAAT – ''Mesorhizobium loti''<br /> | ||
| - | **[[2z9v]] – MlAAT + pyridoxamine<br /> | ||
| - | |||
| - | *Aspartate aminotransferase binary complex with pyridoxal phosphate (PLP) | ||
| - | |||
| - | **[[3ii0]], [[6dnd]] - hAAT + PLP <br /> | ||
| - | **[[6dna]], [[6dnb]] - hAAT (mutant) + PLP <br /> | ||
| - | **[[1asm]], [[1asn]], [[1ars]], [[1art]] - EcAAT + PLP<br /> | ||
| - | **[[3aat]], [[1aam]], [[1aaw]], [[1asf]], [[1asg]], [[1ahe]], [[1ahg]], [[5eaa]], [[1g4v]], [[1g4x]], [[1g7w]], [[1g7x]], [[1ix6]], [[1ix8]], [[2d65]], [[2d66]], [[2d7y]], [[3zzj]], [[3zzk]], [[4a00]] - EcAAT (mutant) + PLP<br /> | ||
| - | **[[7aat]], [[8aat]], [[1tar]], [[1tat]], [[2cst]] - cAAT + PLP<br /> | ||
| - | **[[1asa]], [[1asb]], [[1aka]] - cAAT (mutant) + PLP<br /> | ||
| - | **[[1gc3]], [[1b5o]], [[1b5p]], [[5bj4]] - TtAAT (mutant) + PLP – ''Thermus thermophilus''<br /> | ||
| - | **[[1gd9]] - PhAAT + PLP – ''Pyrococcus horikoshii''<br /> | ||
| - | **[[1j32]] - AAT + PLP – ''Phormidium lapideum''<br /> | ||
| - | **[[1o4s]] - TmAAT + PLP – ''Thermotoga maritima''<br /> | ||
| - | **[[5bj3]] - AAT (mutant) + PLP – ''Thermus aquaticus''<br /> | ||
| - | **[[3k7y]] - AAT + PLP – ''Plasmodium falciparum''<br /> | ||
| - | **[[3ppl]], [[5iwq]] - CgAAT + PLP – ''Corynebacterium glutamicum''<br /> | ||
| - | **[[2z9w]] - MlAAT + pyridoxal<br /> | ||
| - | **[[1daa]], [[4daa]] - BaAAT + PLP – ''Bacillus''<br /> | ||
| - | **[[2dab]], [[5daa]] - BaAAT (mutant) + PLP <br /> | ||
| - | **[[6ezl]] - AAT + PLP – ''Trypanosoma cruzi'' <br /> | ||
| - | |||
| - | *Aspartate aminotransferase binary complex with PLP derivative | ||
| - | |||
| - | **[[1ama]], [[1tas]], [[1oxo]], [[1oxp]], [[1ivr]] - cAAT + PLP derivative<br /> | ||
| - | **[[1akb]], [[1akc]] - cAAT (mutant) + PLP derivative<br /> | ||
| - | **[[1spa]], [[1asl]], [[1asd]], [[1ase]], [[1arg]], [[1c9c]], [[1cq6]], [[1cq7]], [[1cq8]], [[3qn6]], [[3qpg]], [[4f5m]] - EcAAT + PLP derivative<br /> | ||
| - | **[[1asc]], [[1arh]], [[1bqa]], [[1bqd]], [[1toe]], [[4f5f]], [[4f5g]], [[4f5h]], [[4f5i]], [[4f5j]], [[4f5k]], [[4f5l]] - EcAAT (mutant) + PLP derivative<br /> | ||
| - | **[[1x28]], [[1x29]], [[1x2a]] - EcAAT + phosphopyridoxyl glutamate derivative<br /> | ||
| - | **[[1maq]], [[1map]] – cAAT + ketamine<br /> | ||
| - | **[[1ajr]], [[1ajs]], [[5toq]], [[5vk7]] - pAAT + PLP derivative – pig<br /> | ||
| - | **[[5ton]], [[5tor]], [[5tot]] - pAAT (mutant) + PLP derivative<br /> | ||
| - | **[[1bjw]] - TtAAT + PLP derivative<br /> | ||
| - | **[[2gb3]] - TmAAT + PLP derivative<br /> | ||
| - | **[[3meb]] - AAT + PLP derivative – ''Giardia lamblia''<br /> | ||
| - | **[[3hlm]] - mAAT + PLP derivative<br /> | ||
| - | **[[3f6t]] - AAT + PLP derivative – ''Lactobacillus acidophilus''<br /> | ||
| - | **[[4eu1]], [[4w5k]] - AAT + PLP derivative – ''Trypanosoma brucei''<br /> | ||
| - | **[[3nra]] - AAT + PLP derivative – ''Rhodobacter sphaeroides''<br /> | ||
| - | **[[2z9x]] - MlAAT + pyridoxyl-alanine<br /> | ||
| - | **[[1a0g]], [[2daa]], [[3daa]] - BaAAT + PLP derivative <br /> | ||
| - | **[[6f35]] - RmAAT + PLP derivative - ''Rhizobium meliloti''<br /> | ||
| - | |||
| - | *Aspartate aminotransferase ternary complex | ||
| - | |||
| - | **[[1ahf]] - EcAAT (mutant) + indolylpropionic acid + PLP<br /> | ||
| - | **[[1ahx]], [[1tog]] - EcAAT (mutant) + phenylpropionic acid + PLP<br /> | ||
| - | **[[1toi]], [[1toj]] - EcAAT (mutant) + phenylpropionic acid + PLP derivative<br /> | ||
| - | **[[5vwq]], [[5vwr]] - EcAAT + pyridoxal phosphate derivative + inhibitor<br /> | ||
| - | **[[1ahy]], [[1ari]], [[1qir]], [[1qis]], [[1qit]], [[1b4x]], [[1ix7]], [[2d61]], [[2d7z]] - EcAAT (mutant) + maleic acid + PLP<br /> | ||
| - | **[[2q7w]] - EcAAT (mutant) + thiophenecarboxylic acid + PLP<br /> | ||
| - | **[[2qbt]], [[2qa3]], [[2qb2]], [[2qb3]] - EcAAT + thiophenecarboxylic acid + PLP<br /> | ||
| - | **[[1tok]] - EcAAT (mutant) + maleic acid + PLP derivative<br /> | ||
| - | **[[1czc]] - EcAAT (mutant) + glutaric acid + PLP<br /> | ||
| - | **[[1cze]] - EcAAT (mutant) + succinic acid + PLP<br /> | ||
| - | **[[1yoo]], [[2d5y]], [[2d63]], [[2d64]] - EcAAT (mutant) + isovaleric acid + PLP<br /> | ||
| - | **[[3pa9]], [[3paa]] - EcAAT + furancarboxylic acid + pyridoxamine phosphate<br /> | ||
| - | **[[1gc4]], [[1gck]] - TtAAT (mutant) + aspartic acid + PLP<br /> | ||
| - | **[[1gc3]] - TtAAT (mutant) + tryptophan + PLP<br /> | ||
| - | **[[1bkg]] - TtAAT + maleic acid + pyridoxamine phosphate<br /> | ||
| - | **[[1gde]] - PhAAT + glutamic acid + PLP<br /> | ||
| - | **[[1yaa]] - yAAT + maleic acid + PLP – yeast<br /> | ||
| - | **[[3pdb]] - mAAT + oxaloacetic acid + pyridoxamine phosphate<br /> | ||
| - | **[[5vjz]] - pAAT + PLP derivative + methyl-Asp<br /> | ||
| - | **[[6f77]] - RmAAT + PLP derivative + PLP <br /> | ||
| - | **[[5hxx]] - CgAAT + PLP + PLP derivative <br /> | ||
| - | |||
| - | *Bifunctional aspartate aminotransferase | ||
| - | |||
| - | **[[6f5v]], [[5wmh]] - AtBAAT + PLP – ''Arbidopsis thaliana''<br /> | ||
| - | **[[5wmi]], [[5wmk]], [[5wml]] - AtBAAT (mutant) + PLP <br /> | ||
| - | |||
| - | }} | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Han Q, Robinson H, Cai T, Tagle DA, Li J. Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV. Biosci Rep. 2010 Oct 26. PMID:20977429 doi:10.1042/BSR20100117
- ↑ DeLorenzo RJ, Ruddle FH. Glutamate oxalate transaminase (GOT) genetics in Mus musculus: linkage, polymorphism, and phenotypes of the Got-2 and Got-1 loci. Biochem Genet. 1970 Apr;4(2):259-73. PMID:4193185
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Jeffery CJ, Gloss LM, Petsko GA, Ringe D. The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase. Protein Eng. 2000 Feb;13(2):105-12. PMID:10708649
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 Kamitori S, Okamoto A, Hirotsu K, Higuchi T, Kuramitsu S, Kagamiyama H, Matsuura Y, Katsube Y. Three-dimensional structures of aspartate aminotransferase from Escherichia coli and its mutant enzyme at 2.5 A resolution. J Biochem. 1990 Aug;108(2):175-84. PMID:2121725
- ↑ de la Torre F, Cañas RA, Pascual MB, Avila C, Cánovas FM. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J Exp Bot. 2014 Oct;65(19):5527-34. PMID:24902885 doi:10.1093/jxb/eru240
- ↑ Palaiologos G, Hertz L, Schousboe A. Role of aspartate aminotransferase and mitochondrial dicarboxylate transport for release of endogenously and exogenously supplied neurotransmitter in glutamatergic neurons. Neurochem Res. 1989 Apr;14(4):359-66. PMID:2569674
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Tran A, Longo F, Ouzan D, Bianchi D, Pradier C, Saint-Paul MC, Sattonnet C, Laffont C, Dantin S, Piche T, Benzaken S, Rampal P. Effects of 1-year interferon-alpha 2a treatment in patients with chronic hepatitis C and persistently normal transaminase activity. Scand J Gastroenterol. 2000 Apr;35(4):433-7. PMID:10831269
- ↑ 8.0 8.1 8.2 8.3 8.4 Martinez-Carrion M, Tiemeier DC, Peterson DL. Conformational properties of the isoenzymes of aspartate transaminase and the enzyme-substrate complexes. Biochemistry. 1970 Jun 23;9(13):2574-82. PMID:5450225
- ↑ 9.0 9.1 Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J Neurosci. 2000 Dec 15;20(24):8972-9. PMID:11124972
- ↑ 10.0 10.1 Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J Neurosci. 2000 Dec 15;20(24):8972-9. PMID:11124972
- ↑ 11.0 11.1 Jungas RL, Halperin ML, Brosnan JT. Quantitative analysis of amino acid oxidation and related gluconeogenesis in humans. Physiol Rev. 1992 Apr;72(2):419-48. PMID:1557428
- ↑ 12.0 12.1 Gibbs ME, Hertz L. Importance of glutamate-generating metabolic pathways for memory consolidation in chicks. J Neurosci Res. 2005 Jul 15;81(2):293-300. PMID:15929064 doi:10.1002/jnr.20548
- ↑ 13.0 13.1 13.2 13.3 13.4 Gonzalez-Flecha B, Cutrin JC, Boveris A. Time course and mechanism of oxidative stress and tissue damage in rat liver subjected to in vivo ischemia-reperfusion. J Clin Invest. 1993 Feb;91(2):456-64. PMID:8432855 doi:http://dx.doi.org/10.1172/JCI116223