User:Nicholas Bantz/Sandbox 1
From Proteopedia
< User:Nicholas Bantz(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
== Introduction == | == Introduction == | ||
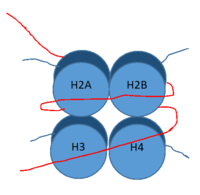
[[Image:Histone.png|200 px|right|thumb|Figure 1: DNA (red) wrapped around histone proteins with histone tails (blue)]] | [[Image:Histone.png|200 px|right|thumb|Figure 1: DNA (red) wrapped around histone proteins with histone tails (blue)]] | ||
| - | <scene name='81/811090/Overall_lsd-1/1'>LSD-1</scene>, human lysine-specific demethylase 1, is an enzyme that affects the ability of DNA to associate with [https://en.wikipedia.org/wiki/Histone histone proteins]. Histone proteins | + | <scene name='81/811090/Overall_lsd-1/1'>LSD-1</scene>, human lysine-specific demethylase 1, is an enzyme that affects the ability of DNA to associate with [https://en.wikipedia.org/wiki/Histone histone proteins]. Histone proteins contain residues that give them an overall positive charge and allow them to act as spools upon which negatively charged DNA can wrap around for storage in the nucleus (Figure 1). When DNA is tightly condensed it forms into nucleosomes which consist of 8 histone core proteins (2 H2A, 2 H2B, 2 H3, 2 H4) with DNA tightly coiled around them. This tightly coiled DNA is known as [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which is inaccessible to transcription factors and RNA polymerase. This can be reversed by modifications to histone protein structure that cause the DNA to relax and form [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which allows for RNA polymerase and other transcription factors to properly execute transcription. One key histone modification is the [https://en.wikipedia.org/wiki/Demethylase demethylation] of lysine residues. Before 2004, it was believed that methylation of histone tails was stable and irreversible. In 2004, it was discovered that histone tails can also be demethylated by demethylase enzymes such as LSD-1 <ref name="Shi">doi: 10.1016/j.cell.2004.12.012</ref>. LSD-1 specifically demethylates mono- or di-methylated lysine substrates at Lys4 or Lys9 in the tail of histone H3. Demethylation of these lysine residues is commonly associated with transcriptional activation, but it also has the ability to silence genes depending on the residue being demethylated, the cofactors present, and the environment in which the demethylation occurs. LSD-1 is among the most well-known demethylases and has been studied since its instrumental discovery in 2004 <ref name="Shi"/>. |
== Structure == | == Structure == | ||
| Line 15: | Line 15: | ||
=== SWIRM Domain === | === SWIRM Domain === | ||
| - | The <scene name='81/811088/Swirmdomain/7'>SWIRM domain</scene> is seen in numerous enzymes that participate in histone binding and chromatin modification. The SWIRM domain of LSD-1 is 94 residues long and is comprised of an alpha-helix bundle <ref name="Stavropolous"/>. The longest helix, 𝛂C, separates the two other helix-turn-helix motifs, <scene name='81/811088/Swirmmotifs/5'>𝛂A/B and 𝛂D/E </scene><ref name="Stavropolous"/>. The SWIRM domain is associated with the oxidase domain via hydrophobic [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between a set of<scene name='81/811088/Oxidaseandswirmchillin/ | + | The <scene name='81/811088/Swirmdomain/7'>SWIRM domain</scene> is seen in numerous enzymes that participate in histone binding and chromatin modification. The SWIRM domain of LSD-1 is 94 residues long and is comprised of an alpha-helix bundle <ref name="Stavropolous"/>. The longest helix, 𝛂C, separates the two other helix-turn-helix motifs, <scene name='81/811088/Swirmmotifs/5'>𝛂A/B and 𝛂D/E </scene><ref name="Stavropolous"/>. The SWIRM domain is associated with the oxidase domain via hydrophobic [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between a set of <scene name='81/811088/Oxidaseandswirmchillin/7'>3 𝛂-helices from each domain</scene>: 𝛂A, 𝛂B, and 𝛂E motifs in the SWIRM domain and 𝛂A, 𝛂B, 𝛂M, motifs in the oxidase domain. The residues that create this hydrophobic interface (which spans nearly 1680 Ų) are practically invariant across histone-modifying proteins <ref name="Stavropolous"/>. The <scene name='81/811090/Hydrophobic_interface_new/5'> hydrophobic interface between the oxidase and SWIRM domains</scene> creates a cleft or tunnel that is also present in other chromatin modifying enzymes. This <scene name='81/811090/Hydrophobic_interface_new/4'>cleft</scene> is responsible for binding to DNA in the other enzymes through the presence of positively charged residues in the cleft <ref name="Stavropolous"/>. The SWIRM domain in LSD-1 is unique because the cleft that is formed by the hydrophobic SWIRM-oxidase interactions lacks the positively charged residues common in other enzymes <ref name="Stavropolous"/>. For this reason, it is proposed that the SWIRM cleft is used for binding the substrate histone tail in order to correctly position the substrate lysine. Multiple experiments showed that mutations in hydrophobic residues that form the SWIRM-oxidase interface greatly reduced the catalytic activity of LSD-1 <ref name="Stavropolous"/>. This, and the proximity to the active site in the oxidase domain, show the importance of the SWIRM cleft in the mechanism of LSD-1. |
=== Oxidase Domain === | === Oxidase Domain === | ||
| - | The <scene name='81/811088/Oxidasedomain/4'>oxidase domain</scene> houses the catalytic site of LSD-1. The domain has two distinct regions: one non-covalently binds the FAD cofactor and the other acts in both the binding and recognition of the substrate lysine on the H3 histone tail<ref name="Stavropolous"/>. The | + | The <scene name='81/811088/Oxidasedomain/4'>oxidase domain</scene> houses the catalytic site of LSD-1. The domain has two distinct regions: one non-covalently binds the FAD cofactor and the other acts in both the binding and recognition of the substrate lysine on the H3 histone tail<ref name="Stavropolous"/>. The substrate-binding subunit of the oxidase domain contains the active site cavity of LSD-1. In relation to other oxidases that utilize FAD as a cofactor, LSD-1 has a very large active site cavity that is 15 Å deep and around 25 Å wide <ref name="Stavropolous"/>. In comparison, [https://en.wikipedia.org/wiki/Polyamine_oxidase polyamine oxidase], another FAD-dependent oxidase, has a catalytic chamber roughly 30 Å long but only a few angstroms wide <ref name=”Binda”>PMID:11258887</ref>. The relatively large size of the LSD-1 active site cavity suggests that other residues, in addition to the substrate lysine, enter into the active site during catalysis. These additional residues could participate in substrate recognition and may contribute to the specificity of LSD-1 for H3K4 and H3K9. |
====Active Site and FAD Cofactor==== | ====Active Site and FAD Cofactor==== | ||
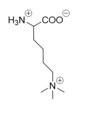
| - | Within the active site cavity, there are four <scene name='81/811090/Active_site_pockets/3'>active site pockets</scene> each with differing chemical properties. The catalytic pocket or invagination within the active site (residues Val317, Gly330, Ala331, Met332, Val333, Phe538, Leu659, Asn660, Lys661, Trp695, Ser749, Ser760 and Tyr761) facilitates the interaction between the FAD cofactor and the substrate lysine <ref name="Stavropolous"/>. This pocket binds and positions the substrate lysine so that it is exposed to the <scene name='81/811089/Fadcofactor/5'>FAD cofactor</scene>. During catalysis, the FAD cofactor is reduced and becomes an anion. Therefore, a positively charged residue is present in most FAD-dependent oxidases to assist in stabilizing the anionic form of FAD. In LSD-1, <scene name='81/811088/Lys661/ | + | Within the active site cavity, there are four <scene name='81/811090/Active_site_pockets/3'>active site pockets</scene> each with differing chemical properties. The catalytic pocket or invagination within the active site (residues Val317, Gly330, Ala331, Met332, Val333, Phe538, Leu659, Asn660, Lys661, Trp695, Ser749, Ser760 and Tyr761) facilitates the interaction between the FAD cofactor and the substrate lysine <ref name="Stavropolous"/>. This pocket binds and positions the substrate lysine so that it is exposed to the <scene name='81/811089/Fadcofactor/5'>FAD cofactor</scene>. During catalysis, the FAD cofactor is reduced and becomes an anion. Therefore, a positively charged residue is present in most FAD-dependent oxidases to assist in stabilizing the anionic form of FAD. In LSD-1, <scene name='81/811088/Lys661/5'>Lys661</scene> is present in the catalytic pocket of the active site to help stabilize the negatively charged FAD <ref name="Stavropolous"/>. The other three <scene name='81/811090/Active_site_pockets/3'>pockets</scene> are not as well understood but predictions can be made about their functions within the active site of LSD-1. Because the active site is able to accept additional residues on the substrate histone accompanying the lysine, the remaining three pockets are most plausibly responsible for the recognition of regions with distinct chemical properties on the histone tail<ref name="Stavropolous"/>. The first pocket (Pocket 1) that assists in recognizing chemical modifications on the substrate histone is composed of residues Val334, Thr335, Asn340, Met342, Tyr571, Thr810, Val811 and His812 <ref name="Stavropolous"/>. The second pocket in the active site for side-chain recognition (Pocket 2) is composed of Phe558, Glu559, Phe560, Asn806, Tyr807 and Pro808 <ref name="Stavropolous"/>. Pocket 3 within the active site is composed of Asn540, Leu547, Trp552, Asp553, Gln554, Asp555, Asp556, Ser762, Tyr763, Val764 and Tyr773 <ref name="Stavropolous"/>. Each of the three pockets, in addition to the catalytic pocket, are able to recognize distinct chemical properties on the substrate and contribute to the specificity of LSD-1. |
| Line 28: | Line 28: | ||
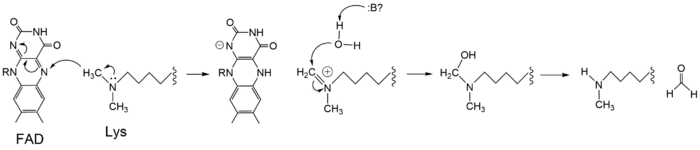
| - | The mechanism of lysine demethylation is highly dependent on the presence of the <scene name='81/811089/Fadcofactor/5'>FAD cofactor</scene>. The FAD cofactor, positioned closely to the substrate lysine in the active site, acts as an oxidizing agent and initiates catalysis (Figure 3). A two-electron transfer occurs between the substrate lysine and the isoallozaxine ring of the FAD cofactor in the form of a [https://en.wikipedia.org/wiki/Hydride hydride]; the substrate lysine is oxidized and the FAD is reduced <ref name="Stavropolous"/>. The FAD cofactor forms an anion and is stabilized by the positively charged <scene name='81/811088/Lys661/ | + | The mechanism of lysine demethylation is highly dependent on the presence of the <scene name='81/811089/Fadcofactor/5'>FAD cofactor</scene>. The FAD cofactor, positioned closely to the substrate lysine in the active site, acts as an oxidizing agent and initiates catalysis (Figure 3). A two-electron transfer occurs between the substrate lysine and the isoallozaxine ring of the FAD cofactor in the form of a [https://en.wikipedia.org/wiki/Hydride hydride]; the substrate lysine is oxidized and the FAD is reduced <ref name="Stavropolous"/>. The FAD cofactor forms an anion and is stabilized by the positively charged <scene name='81/811088/Lys661/5'>Lys661</scene> positioned in the catalytic pocket of the active site <ref name="Stavropolous"/>. The oxidized substrate lysine forms an iminium cation that is hydrolyzed into the carbinolamine intermediate <ref name="Stavropolous"/>. The carbinolamine intermediate readily decomposes into formaldehyde and the demethylated lysine substrate <ref name="Stavropolous"/>. |
===Inhibition by Tri-Methylated Lysine=== | ===Inhibition by Tri-Methylated Lysine=== | ||
| Line 41: | Line 41: | ||
===2. Cancer=== | ===2. Cancer=== | ||
| - | Research has found that LSD-1 is over-expressed in many tumorous cancers. The proposed mechanism behind the carcinogenic role of LSD-1 focuses on the known tumor-suppressor gene, [https://en.wikipedia.org/wiki/P53 p53]. The p53 protein acts as a transcription factor that activates the expression of many anti-proliferative proteins. LSD-1 has been found to remove a methyl group from the di-methylated Lys370 on p53 <ref name="Jin"> | + | Research has found that LSD-1 is over-expressed in many tumorous cancers. The proposed mechanism behind the carcinogenic role of LSD-1 focuses on the known tumor-suppressor gene, [https://en.wikipedia.org/wiki/P53 p53]. The p53 protein acts as a transcription factor that activates the expression of many anti-proliferative proteins. LSD-1 has been found to remove a methyl group from the di-methylated Lys370 on p53 <ref name="Jin">doi: 10.1042/BJ20121360</ref>. Similar to the proposed role of LSD-1 in diabetes, its demethylation of p53 is inhibitory and prevents its binding to DNA <ref name="Jin"/>. This inactivation of p53 is thought to prevent anti-proliferative operations in the cell and contribute to the development of multiple types of cancers. |
== References == | == References == | ||
Current revision
LSD-1: Human lysine-specific demethylase 1
| |||||||||||
Student Contributors
- Nicholas Bantz
- Cody Carley
- Michael Thomas