User:Emily Leiderman/Sandbox 1
From Proteopedia
< User:Emily Leiderman(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 32: | Line 32: | ||
== Mechanism == | == Mechanism == | ||
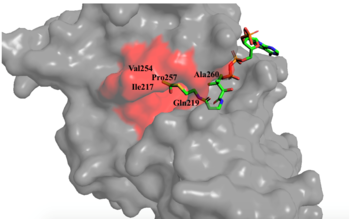
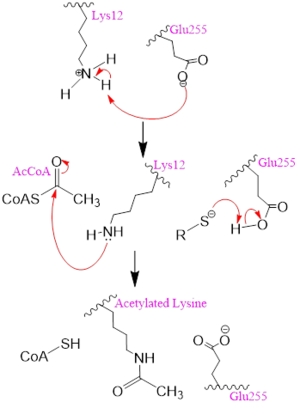
| - | [[Image: | + | [[Image:mechh.PNG|300px|left|thumb|Figure 2. A possible acetylation mechanism of HAT1 (PDB: 1BOB). Mechanism consists of deptronation of Lys-12 of histone H4 and then transfer of an acetyl group from Acetyl CoA to Lys-12.]] Several mechanisms for the transfer of the acetyl group to the Lys-12 of the histone have been proposed. The most accurate and likely mechanism today is conserved across several HAT1 studies and consists of deprotonating the amino group on the Lys-12 of H4 (figure 2). This mechanism was proposed by Wu et al with respect to the human HAT1 protein. Analysis was done on the 1BOB model (retrieved from the protein database) and similar residues with respect to location and function were identified. The main chain carbonyl oxygen of the polar residues <scene name='81/811098/Gln-219_glu-255_asp-256/3'>Gln-219, Glu-255, and Asp-256</scene> are predicted to deprotonate the Lys-12 reside of H4 as seen in figure 2 <ref name=Wu>PMID:22615379 </ref>. Structural analysis of the 4PSW protein shows that the main chain carbonyls of these three residues are found in close proximity of the amino group of Lys-12 <ref name=idk>PMID: 24835250 </ref>. Of those residues it is uncertain which (if any) accept the proton of Lys-12. Each main chain carbonyl oxygen belonging to these three residues falls within 2 to 3 angstroms of the target amide on Lysine 12. Deprotonation of the amino group on the Lys-12 makes the residue nucleophilic enough to directly attack the carbonyl carbon of the acetyl group to initiate the acetyl transfer. We selected Glu255 to be the proton acceptor in our proposed mechanism displayed in figure 2. The transfer mechanism is contingent on the conformational change and the formation of a functional gate that spans the concave groove over the bound Acetyl CoA. This gate holds the substrate in place while the enzymatic [https://en.wikipedia.org/wiki/Deprotonation deprotonation] process takes place <ref name=Wu/>. |
| Line 38: | Line 38: | ||
=== Protein Gate === | === Protein Gate === | ||
| - | The <scene name='81/811098/Gate/5'>protein gate</scene> consists of several residues that coordinate a conformational change in the protein to hold the Acetyl CoA ligand in place. <scene name='81/811098/161_162_258/1'> | + | The <scene name='81/811098/Gate/5'>protein gate</scene> consists of several residues that coordinate a conformational change in the protein to hold the Acetyl CoA ligand in place. <scene name='81/811098/161_162_258/1'>Asn-258, Glu-162, Ile-161</scene> cooperate to create a bridge over Acetyl CoA <ref name=Dut/>. The main chain amide of <scene name='81/811098/Phe-261_asn-258/1'>Phe-261</scene> hydrogen bonds to the carbonyl oxygen of Asn-258 which allows for proper positioning of Asn-258 to bond with other residues and Acetyl CoA itself. The side chain amide of Asn-258 also binds via a water molecule (H2O-415) to Glu-162 on its main chain amide <ref name=Dut/>. These bonds help secure the position of <scene name='81/811098/Asn-258/4'>Asn-258</scene> so that a hydrogen bond can be established from the side-chain amide to the PO5 carbonyl oxygen. Together, the side chains of Ile-161, Glu-162, and Asn-258 form a protein gate over the Acetyl CoA binding cleft. Hydrogen bonds further fixate Acetyl CoA by connecting the main chain amide of Glu-162 to the side chain amide of Asn-258. The side chain carbonyl oxygen of Phe-261 hydrogen bonds to its main chain amide to bridge the binding cleft over Acetyl-CoA. The gate allows for correct binding of Acetyl CoA. Once the ligand is bound, subsequent conformational changes of HAT1 take place allowing for Lys-12 to act as a nucleophile. |
| + | |||
== Medical Relevance == | == Medical Relevance == | ||
| + | Histone acetyltransferases consists of 5 families: GNAT, MYST, p300/CBP, and Rtt109. HATs of these families have histone substrate specificity, such as lysine, and non-histone substrate specificity, such as proteins and transcription factors. Specifically, the acetylation of the particular residue, lysine 16 of histone H4, is crucial in maintaining a normal [https://en.wikipedia.org/wiki/LMNA lamin A] gene and genomic stability since a mutation in the lamin A gene can cause premature, rapid aging <ref name=idkk>PMID:21746928 </ref>. [https://en.wikipedia.org/wiki/progeria Hutchinson Gilford progeria], an example of an accelerated aging syndrome, is caused by a mutated lamin A producing the mutant protein [https://en.wikipedia.org/wiki/Progerin progerin], resulting in the delayed response time of [https://en.wikipedia.org/wiki/DNA_repair repair proteins] during DNA damage <ref name=idkk/>. A study done by Krishnan et. al showed that it was a defect in histone acetyltransferase, Mof, in [https://en.wikipedia.org/wiki/ZMPSTE24 Zmpste24-/-] mice that allowed for the hypoacetylation of lysine 16 residue (H4K16) which lead to a delayed response in modified histones recruiting repair proteins to fix DNA damage. Thus, the inability of repair proteins to correct DNA damage suggests that cells get inclined to rapid aging and early [https://en.wikipedia.org/wiki/Senescence senescence], resulting in syndromes such as Hutchinson Gilford progeria <ref name=idkk/>. | ||
Current revision
Histone Acetyltransferase 1
| |||||||||||