We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sean Callahan/Sandbox 1
From Proteopedia
< User:Sean Callahan(Difference between revisions)
| Line 10: | Line 10: | ||
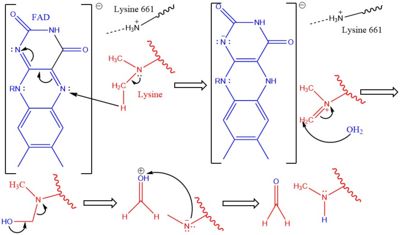
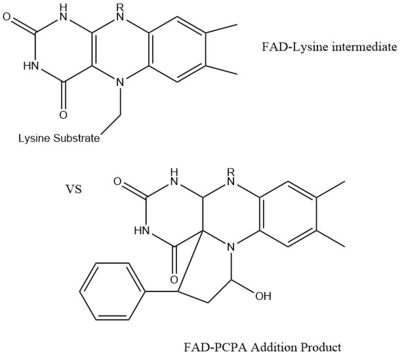
LSD1 is found in ''Homo sapiens'' and its crystal structure was obtained in the presence of the FAD cofactor. LSD1, when compared to other histone binding proteins, contains both conserved and novel structural features. Its substrates are mono- or di-methylated lysine residues and its products are demethylated or mono-methylated lysine residues, respectively. | LSD1 is found in ''Homo sapiens'' and its crystal structure was obtained in the presence of the FAD cofactor. LSD1, when compared to other histone binding proteins, contains both conserved and novel structural features. Its substrates are mono- or di-methylated lysine residues and its products are demethylated or mono-methylated lysine residues, respectively. | ||
===N-Terminus=== | ===N-Terminus=== | ||
| - | Going in order of primary structure, the first 166 residues are believed to be unstructured and contain a nuclear localization signal. This area of the protein has also been shown to be susceptible to proteolytic cleavage, which may be to remove the localization signal and render protein inactive<ref name="Stavropoulos">PMID: 16799558</ref>. However, a mutant of LSD1, which contains residues 166-852 (essentially eliminating the unstructured region) has been shown to be stable and viable when compared to wild-type LSD1 in a photometric activity assay<ref name="Stavropoulos">PMID: 16799558</ref>. Unfortunately, this portion of the protein was unable to be crystallized<ref name="Stavropoulos">PMID: 16799558</ref>. | + | Going in order of primary structure, the first 166 residues are believed to be unstructured and contain a nuclear localization signal. This area of the protein has also been shown to be susceptible to proteolytic cleavage, which may be to remove the localization signal and render protein inactive<ref name="Stavropoulos">PMID: 16799558</ref>. However, a mutant of LSD1, which contains residues 166-852 (essentially eliminating the unstructured region) has been shown to be stable and viable when compared to wild-type LSD1 in a photometric activity assay <ref name="Stavropoulos">PMID: 16799558</ref>. Unfortunately, this portion of the protein was unable to be crystallized<ref name="Stavropoulos">PMID: 16799558</ref>. |
===SWIRM Domain=== | ===SWIRM Domain=== | ||
The next section of LSD1 spans residues 166-260 and is called the <scene name='81/811711/Swirm_domain/4'>SWIRM Domain</scene>, named after the [https://www.pnas.org/content/pnas/103/7/2057.full.pdf?with-ds=yes SWI3, RSC8, and MOIRA] proteins from which it was first discovered. It is a conserved domain among histone binding proteins, however LSD1's SWIRM domain is unique in that it does not have a positively charged DNA binding domain on the exterior of the protein. Because of this, it believed that LSD1 does not directly bind DNA unlike other histone binding proteins <ref name="Da">PMID: 16461455</ref>. The conserved secondary structure of this domain is characterized by a long central helix, with two, shorter helix motifs surrounding it. | The next section of LSD1 spans residues 166-260 and is called the <scene name='81/811711/Swirm_domain/4'>SWIRM Domain</scene>, named after the [https://www.pnas.org/content/pnas/103/7/2057.full.pdf?with-ds=yes SWI3, RSC8, and MOIRA] proteins from which it was first discovered. It is a conserved domain among histone binding proteins, however LSD1's SWIRM domain is unique in that it does not have a positively charged DNA binding domain on the exterior of the protein. Because of this, it believed that LSD1 does not directly bind DNA unlike other histone binding proteins <ref name="Da">PMID: 16461455</ref>. The conserved secondary structure of this domain is characterized by a long central helix, with two, shorter helix motifs surrounding it. | ||
| Line 22: | Line 22: | ||
==Regulation== | ==Regulation== | ||
=== Allosteric Stie === | === Allosteric Stie === | ||
| - | LSD-1 can be regulated by outside factors. The tower domain and oxidase domain are connected by a <scene name='81/811711/Regulator_loop/7'>tower-oxidase connector</scene>, also referred to as the regulator loop in literature. The oxidase domain is what harbors the catalytic chamber for LSD-1. Any change to the catalytic chamber could drastically reduce the ability for the methylated lysine of the substrate to fit in the binding site correctly. Because the tower domain is attached to the oxidase domain, the tower domain and the connector region may allosterically regulate demethylase activity. Any interaction with these two sites is hypothesized to change the enzyme activity. This has further pushed the assumption that the CoRest complex effects enzyme activity via interaction with the tower domain | + | LSD-1 can be regulated by outside factors. The tower domain and oxidase domain are connected by a <scene name='81/811711/Regulator_loop/7'>tower-oxidase connector</scene>, also referred to as the regulator loop in literature. The oxidase domain is what harbors the catalytic chamber for LSD-1 <ref name="Stavropoulos">PMID: 16799558</ref>. Any change to the catalytic chamber could drastically reduce the ability for the methylated lysine of the substrate to fit in the binding site correctly. Because the tower domain is attached to the oxidase domain, the tower domain and the connector region may allosterically regulate demethylase activity. Any interaction with these two sites is hypothesized to change the enzyme activity. This has further pushed the assumption that the CoRest complex effects enzyme activity via interaction with the tower domain <ref name="Stavropoulos">PMID: 16799558</ref>. |
===Androgen Receptor=== | ===Androgen Receptor=== | ||
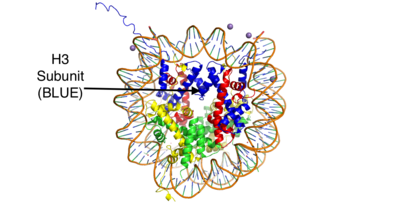
LSD-1 will demethylate mono or di-methylated lysines. It will only demethylate lysine 4 of histone 3. This factor can be regulated by the androgen receptor which can change its specificity for a different target. The [http://proteopedia.org/wiki/index.php/Androgen_receptor#Function androgen receptor] is a steroid hormone receptor that can modulate transcription. When it interacts with LSD-1 it will no longer demethylate H3-K4, but will now demethylate H3-K9. This attribute allows LSD-1 to work on a wider range of residues. | LSD-1 will demethylate mono or di-methylated lysines. It will only demethylate lysine 4 of histone 3. This factor can be regulated by the androgen receptor which can change its specificity for a different target. The [http://proteopedia.org/wiki/index.php/Androgen_receptor#Function androgen receptor] is a steroid hormone receptor that can modulate transcription. When it interacts with LSD-1 it will no longer demethylate H3-K4, but will now demethylate H3-K9. This attribute allows LSD-1 to work on a wider range of residues. | ||
Current revision
Lysine Specific Demethylase 1 (Homo sapiens)
| |||||||||||