User:Lauryn Padgett/Sandbox 1
From Proteopedia
< User:Lauryn Padgett(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 33: | Line 33: | ||
==Inhibitors== | ==Inhibitors== | ||
| - | SET7/9’s structure and function has been studied extensively because of its role in transcription <ref name="Takemoto">PMID:27088648</ref>. In the past few years it has been identified to methylate genes involved in multiple diseases; making it a potential candidate for drug inhibition | + | SET7/9’s structure and function has been studied extensively because of its role in transcription <ref name="Takemoto">PMID:27088648</ref>. In the past few years it has been identified to methylate genes involved in multiple diseases; making it a potential candidate for drug inhibition. <ref name="Tamura">PMID:29723250</ref> Two compounds that have been found to inhibit SET7/9 in certain cells in vitro are Sinefungin and Cyproheptadine. Each inhibitor acts on the catalytic center of SET7/9, however their mechanisms of inhibition and possible medical relevancies differ greatly. |
===Sinefungin=== | ===Sinefungin=== | ||
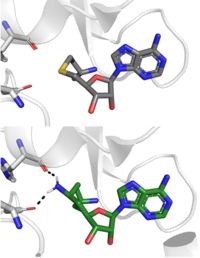
| - | Sinefungin is a potent methyltransferase inhibitor that is a natural nucleoside isolated from the [https://www.britannica.com/science/Streptomyces "Streptomyces"] species. <ref name= | + | Sinefungin is a potent methyltransferase inhibitor that is a natural nucleoside isolated from the [https://www.britannica.com/science/Streptomyces "Streptomyces"] species. <ref name="Tamura" /> Also referred to as adenosyl-ornithine, it is the delta (5’ adenosyl) derivative of [https://en.wikipedia.org/wiki/Ornithine ornithine] and a [https://en.wikipedia.org/wiki/Structural_analog structural analog] of S-adenosylmethionine. Sinefugin is unique because it inhibits where the cofactor binds rather than where the substrate binds like a typical competitive inhibitor. Sinefungin is more stable bound in the active site than SAH due to the ability to create two additional hydrogen bonds to its amine group that are not possible with SAH’s sulfur. |
[[Image: SinSAH.jpg|200 px| right| thumb|SAH (grey) and Sinefungin (green) in the peptide binding pocket. The nitrogen group of sinefungin makes 2 double bonds to the main chain carbonyls of Arg265 and His293. Sinefungin was created using PDB: 1O9S and mutating the sulfur of SAH]] | [[Image: SinSAH.jpg|200 px| right| thumb|SAH (grey) and Sinefungin (green) in the peptide binding pocket. The nitrogen group of sinefungin makes 2 double bonds to the main chain carbonyls of Arg265 and His293. Sinefungin was created using PDB: 1O9S and mutating the sulfur of SAH]] | ||
| - | Sinefungin has been used experimentally to inhibit the SET 7/9 protein on [https://www.sciencedirect.com/topics/medicine-and-dentistry/peritoneal-fibrosis peritoneal fibrosis] in mice and in human peritoneal mesothelial cells. <ref name= | + | Sinefungin has been used experimentally to inhibit the SET 7/9 protein on [https://www.sciencedirect.com/topics/medicine-and-dentistry/peritoneal-fibrosis peritoneal fibrosis] in mice and in human peritoneal mesothelial cells. <ref name="Tamura" /> SET 7/9 is involved in peritoneal fibrosis because it mono-methylates [https://epigenie.com/key-epigenetic-players/histone-proteins-and-modifications/histone-h3k4/ H3K4], which activates the transcription of fibrosis related genes. The administration of Sinefungin to mice in vitro resulted in decreased levels of methylated H3K4 (H3K4me1) protein, as well as suppressed peritoneal cell density and thickening. The decreased levels of H3K4me1 suggest that the methylation of H3K4 was inhibited by Sinefungin, as well as that inhibiting SET7/9 ameliorates peritoneal fibrosis. |
Current revision
Histone Lysine Methyltransferase: Gene Activator
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1016/j.apsb.2013.04.007
- ↑ 3.0 3.1 Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. PMID:23566087 doi:http://dx.doi.org/10.2217/epi.13.13
- ↑ 4.0 4.1 Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011 Sep 1;6(9):1059-67. doi: 10.4161/epi.6.9.16069. Epub 2011 Sep, 1. PMID:21847010 doi:http://dx.doi.org/10.4161/epi.6.9.16069
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 6.0 6.1 Takemoto Y, Ito A, Niwa H, Okamura M, Fujiwara T, Hirano T, Handa N, Umehara T, Sonoda T, Ogawa K, Tariq M, Nishino N, Dan S, Kagechika H, Yamori T, Yokoyama S, Yoshida M. Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription. J Med Chem. 2016 Apr 28;59(8):3650-60. doi: 10.1021/acs.jmedchem.5b01732. Epub, 2016 Apr 18. PMID:27088648 doi:http://dx.doi.org/10.1021/acs.jmedchem.5b01732
- ↑ 7.0 7.1 7.2 Tamura R, Doi S, Nakashima A, Sasaki K, Maeda K, Ueno T, Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One. 2018 May 3;13(5):e0196844. doi: 10.1371/journal.pone.0196844., eCollection 2018. PMID:29723250 doi:http://dx.doi.org/10.1371/journal.pone.0196844
Student Contributors
Lauryn Padgett, Alexandra Pentala, Madeleine Wilson