User:Tereza Čalounová/Sandbox 1
From Proteopedia
m |
m |
||
| (26 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | This is | + | This page is a project made with my classmates Kateřina Krausová and Dominik Merta for Structural Biology of the Cell Class in summer 2019 at the Charles University. |
| - | + | ||
| - | + | ||
==Tafazzin== | ==Tafazzin== | ||
{{Theoretical_model}} | {{Theoretical_model}} | ||
| - | Tafazzin is a protein located in mitochondrial inner membranes. It is involved in altering cardiolipin. Cardiolipin is key in maintaining mitochondrial shape, energy production, and protein transport within cells. The full-length tafazzin protein contains 292 amino acids and has a molecular weight of 33459 daltons. | + | Tafazzin is a protein located in mitochondrial inner membranes. It is involved in altering [https://en.wikipedia.org/wiki/Cardiolipin cardiolipin]. Cardiolipin is key in maintaining mitochondrial shape, energy production, and protein transport within cells. The full-length tafazzin protein contains 292 amino acids and has a molecular weight of 33459 daltons. |
| - | Mutations in gene associated with this protein can cause Barth Syndrome. Barth syndrome (BTHS) | + | Mutations in gene associated with this protein can cause Barth Syndrome. Barth syndrome (BTHS) is a genetic disorder diagnosed almost exclusively in males. BTHS is rare, it is estimated to affect 1 in 300,000 to 400,000 individuals worldwide. Males with BTHS have weak heart and skeletal muscles which can lead to heart failure. Another of the symptoms is [https://en.wikipedia.org/wiki/Neutropenia neutropenia] which can lead to infections. <ref name="cit9">Barth syndrome. In: Ghr.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2019 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/condition/barth-syndrome#</ref> |
<StructureSection load='Tafazzinpredictedstructure.pdb' size='350' side='right' caption='Theoretical model of Tafazzin made using [https://swissmodel.expasy.org/repository/uniprot/Q16635 SWISS-MODEL].' scene='81/813423/Tafazzinpredictedcartoon/1'> | <StructureSection load='Tafazzinpredictedstructure.pdb' size='350' side='right' caption='Theoretical model of Tafazzin made using [https://swissmodel.expasy.org/repository/uniprot/Q16635 SWISS-MODEL].' scene='81/813423/Tafazzinpredictedcartoon/1'> | ||
== Function == | == Function == | ||
| Line 12: | Line 10: | ||
*1-acylglycerol-3-phosphate O-acyltransferase activity | *1-acylglycerol-3-phosphate O-acyltransferase activity | ||
*1-acylglycerophosphocholine O-acyltransferase activity | *1-acylglycerophosphocholine O-acyltransferase activity | ||
| - | *O-acyltransferase activity <ref name="cit5">https://www.uniprot.org/uniprot/Q16635?fbclid=IwAR3v10lUTRZfb0NFOYKC4wjaherdU9PIVJ8T63jkC9RfNu_5OQ2IpoDR0iY</ref> | + | *O-acyltransferase activity <ref name="cit5">Q16635 (TAZ_HUMAN). In: Https://www.uniprot.org/ [online]. Cambridge, Geneva, Washington: UniProt, 2019 [cit. 2019-04-27]. Available at: https://www.uniprot.org/uniprot/Q16635?fbclid=IwAR3v10lUTRZfb0NFOYKC4wjaherdU9PIVJ8T63jkC9RfNu_5OQ2IpoDR0iY</ref> |
==== Biological process ==== | ==== Biological process ==== | ||
*cardiac muscle contraction | *cardiac muscle contraction | ||
| Line 22: | Line 20: | ||
*hemopoiesis | *hemopoiesis | ||
*inner mitochondrial membrane organization | *inner mitochondrial membrane organization | ||
| - | *mitochondrial ATP synthesis coupled electron transport | + | *mitochondrial [https://en.wikipedia.org/wiki/Adenosine_triphosphate ATP] synthesis coupled electron transport |
*mitochondrial respiratory chain complex I assembly | *mitochondrial respiratory chain complex I assembly | ||
*muscle contraction | *muscle contraction | ||
| - | *positive regulation of ATP biosynthetic | + | *positive regulation of ATP biosynthetic process |
*positive regulation of cardiolipin metabolic process | *positive regulation of cardiolipin metabolic process | ||
*regulation of gene expression | *regulation of gene expression | ||
| Line 31: | Line 29: | ||
=== Cardiolipin === | === Cardiolipin === | ||
| - | Cardiolipin is a trivial name used for 1,3-bis(sn-3’-phosphatidyl)-sn-glycerol. <ref name="cit12">http://www.lipidhome.co.uk/lipids/complex/dpg/index.htm</ref> It is a phospholipid mostly found in the inner membrane of mitochondria where is essential for stability of enzymes which are part of the energy metabolism. Cardiolipin is also involved in different stages of the mitochondrial apoptotic process and in mitochondrial membrane dynamics. <ref name="cit13"> | + | Cardiolipin is a trivial name used for 1,3-bis(sn-3’-phosphatidyl)-sn-glycerol. <ref name="cit12">CHRISTIE, William. Cardiolipin (Diphosphatidylglycerol). In: Www.lipidhome.co.uk [online]. Hutton: The LipidWeb, 2019 [cit. 2019-04-27]. Available at: http://www.lipidhome.co.uk/lipids/complex/dpg/index.htm</ref> It is a [https://en.wikipedia.org/wiki/Phospholipid phospholipid] mostly found in the inner membrane of mitochondria where is essential for stability of enzymes which are part of the energy metabolism. Cardiolipin is also involved in different stages of the mitochondrial [https://en.wikipedia.org/wiki/Apoptosis apoptotic] process and in mitochondrial membrane dynamics. <ref name="cit13">Houtkooper, R. H., & Vaz, F. M. (2008). Cardiolipin, the heart of mitochondrial metabolism. Cellular and Molecular Life Sciences, 65(16), 2493–2506. doi:10.1007/s00018-008-8030-5</ref> |
==== Structure ==== | ==== Structure ==== | ||
| - | [[Image:CardiolipinStructure.png|thumb|Chemical structure of cardiolipin where R1-R4 are alkyl groups, typically 18-carbon fatty acid side chains. <ref name="cit14">https://commons.wikimedia.org/wiki/File:Cardiolipin_structure.svg?uselang=cs</ref>]] | + | [[Image:CardiolipinStructure.png|thumb|Chemical structure of cardiolipin where R1-R4 are alkyl groups, typically 18-carbon fatty acid side chains. <ref name="cit14">Cardiolipin structure. In: Commons.wikimedia.org [online]. San Francisco: Wikimedia, 2019 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Cardiolipin_structure.svg?uselang=cs</ref>]] |
Cardiolipin’s structure differs from other phospholipids since it is a dimer, it results in a highly specific conical structure. Cardiolipin consists of two phosphatidic acids, three glycerol backbones and four fatty acyl groups, so it can potentially carry two negative charges.<ref name="cit12" /> <ref name="cit13" /> | Cardiolipin’s structure differs from other phospholipids since it is a dimer, it results in a highly specific conical structure. Cardiolipin consists of two phosphatidic acids, three glycerol backbones and four fatty acyl groups, so it can potentially carry two negative charges.<ref name="cit12" /> <ref name="cit13" /> | ||
| - | The composition of fatty acids depends on | + | The composition of fatty acids depends on the organism. With a few exceptions like testis or central nervous system, linoleic acid creates 80 % of fatty acids in animal tissues. Testis contain mainly palmitic acid in contrast to central nervous system which contains a wide range of different fatty acids like palmitic, stearic, oleic and many others.<ref name="cit12" /> |
==== Synthesis ==== | ==== Synthesis ==== | ||
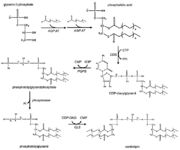
| - | [[Image:CardiolipinSynthesis.jpg|thumb|Cardiolipin synthesis. <ref name="cit16">https://commons.wikimedia.org/wiki/File:Eukaryotic_pathway.jpg</ref>]] | + | [[Image:CardiolipinSynthesis.jpg|thumb|Cardiolipin synthesis. <ref name="cit16"> Rochellehx. File:Eukaryotic pathway.jpg. In: Wikimedia Commons [online]. 23. 4. 2009 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Eukaryotic_pathway.jpg</ref>]] |
| - | The first step in the cardiolipin synthesis is a conversion of cytidine diphosphate diacylglycerol to phosphatidylglycerol phosphate by the enzyme phosphatidylglycerol phosphate (PGP) synthase. | + | The first step in the cardiolipin synthesis is a conversion of cytidine diphosphate diacylglycerol to phosphatidylglycerol phosphate by the enzyme phosphatidylglycerol phosphate (PGP) synthase. It is followed by dephosphorylation by PGP phosphatase to phosphatidylglycerol. Lastly the phosphatidylglycerol is converted to cardiolipin in a condensation reaction catalyzed by cardiolipin synthase. <ref name="cit15">PMID: 24445246</ref> |
| - | The efficiency of these synthesis depends on the amount of phosphatidyl glycerol in the mitochondrial membrane. The lesser amount the bigger effectiveness. <ref name="cit13" /> | + | The efficiency of these synthesis depends on the amount of phosphatidyl glycerol in the mitochondrial membrane. The lesser the amount the bigger the effectiveness. <ref name="cit13" /> |
==== Remodeling ==== | ==== Remodeling ==== | ||
[[Image:RemodelingCardiolipin.png|thumb|Cardiolipin synthesis and remodeling.]] | [[Image:RemodelingCardiolipin.png|thumb|Cardiolipin synthesis and remodeling.]] | ||
| - | The remodeling is based on unsaturation of the acyl chains of the cardiolipin. This process has two steps. Firstly, the fatty acyl group is removed by a specific cardiolipin deacylase forming monolysocardiolipin. Then tafazzin | + | The remodeling is based on unsaturation of the acyl chains of the cardiolipin. This process has two steps. Firstly, the fatty acyl group is removed by a specific cardiolipin deacylase forming monolysocardiolipin. Then tafazzin recycles monolysocardiolipin and makes the final product, unsaturated cardiolipin. <ref name="cit15" /> |
==== Function ==== | ==== Function ==== | ||
| - | As mentioned, cardiolipin is mainly located on the mitochondrial inner membrane, but is also present on the outer mitochondrial membrane. Cardiolipin is essential for some enzyme systems such as energy metabolism, membrane transport and cell division. For example, when the apoptosis, programmed cell death, or mitophagy, mitochondrial degradation, is needed, cardiolipin is externalized on the surface of mitochondria and acts like a signaling molecule. <ref name="cit12" /> | + | As mentioned, cardiolipin is mainly located on the mitochondrial inner membrane, but is also present on the outer mitochondrial membrane. Cardiolipin is essential for some enzyme systems such as energy metabolism, membrane transport and cell division. For example, when the apoptosis, programmed cell death, or [https://en.wikipedia.org/wiki/Mitophagy mitophagy], mitochondrial degradation, is needed, cardiolipin is externalized on the surface of mitochondria and acts like a signaling molecule. <ref name="cit12" /> |
On top of that, cardiolipin has many other functions. It is an important cofactor for cholesterol translocation from the outer to the inner mitochondrial membrane. It also has a role in the regulation of gene expression. What is more, it can act like a molecular chaperone, so it helps mitochondrial proteins with folding. <ref name="cit13" /> | On top of that, cardiolipin has many other functions. It is an important cofactor for cholesterol translocation from the outer to the inner mitochondrial membrane. It also has a role in the regulation of gene expression. What is more, it can act like a molecular chaperone, so it helps mitochondrial proteins with folding. <ref name="cit13" /> | ||
| Line 56: | Line 54: | ||
== TAZ gene == | == TAZ gene == | ||
| - | Tafazzin is encoded by a TAZ gene (G4.5). This gene is located on the long arm of the X chromosome at position 28 (Xq28). <ref name="cit6">https://ghr.nlm.nih.gov/gene/TAZ# | + | Tafazzin is encoded by a TAZ gene (G4.5). This gene is located on the long arm of the X chromosome at position 28 (Xq28). <ref name="cit6">TAZ gene. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2014 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/gene/TAZ#</ref> |
| - | Several isoforms are known. Isoforms that lack the N-terminus are found in leukocytes and fibroblasts, but not in heart and skeletal muscle. <ref name=" | + | Several [https://en.wikipedia.org/wiki/Protein_isoform isoforms] are known. Isoforms that lack the N-terminus are found in [https://en.wikipedia.org/wiki/White_blood_cell leukocytes] and [https://en.wikipedia.org/wiki/Fibroblast fibroblasts], but not in heart and skeletal muscle. <ref name="cit5" /> |
| - | + | Human TAZ consists of 11 [https://en.wikipedia.org/wiki/Exon exons] and produces four alternatively spliced [https://en.wikipedia.org/wiki/Messenger_RNA mRNA] transcripts: a full-length transcript containing all exons (FL), a transcript lacking exon 5 (Δ5), a transcript lacking exon 7 (Δ7) and a transcript lacking both exons 5 and 7 (Δ5Δ7). Of these four alternatively spliced variants of human TAZ, FL and Δ5 encode proteins with transacylase activity, while Δ7 and Δ5Δ7 do not. <ref name="cit8">PMID: 25941633</ref> | |
| - | + | ||
| - | + | ||
| - | + | == Disease - Barth Syndrome== | |
| - | + | Barth syndrome (BTHS), also known as 3-Methylglutaconic aciduria type II, is an X-linked genetic disorder. The disease is caused by mutation in TAZ gene which encodes for protein tafazzin. <ref name="cit9" /> Tafazzin works as an [https://en.wikipedia.org/wiki/Acyltransferase acyltransferase]in complex lipid metabolism, it is responsible for altering immature [https://en.wikipedia.org/wiki/Cardiolipin cardiolipin]- intermediate [https://en.wikipedia.org/wiki/Monolysocardiolipin monolysocardiolipin](with three linoleic acid side chains) (MLCL). <ref name="cit2">Barth syndrome cells display widespread remodeling of mitochondrial complexes without affecting metabolic flux distribution. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease [online]. 2018, 1864(11), 3650-3658 [cit. 2019-04-27]. Available at: https://www.sciencedirect.com/science/article/pii/S092544391830334X?via%3Dihub</ref> <ref name="cit3">Barth syndrome: an X‐linked cause of fetal cardiomyopathy and stillbirth. Prenatal Diagnosis [online]. 2010, 30(10), 970-976 [cit. 2019-04-27]. Available at: https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.2599</ref> Cardiolipin makes up 20% of mitochondrial lipids and is closely connected with the electron transport chain proteins and the inner membrane structure of the mitochondria. <ref name="cit4">Tafazzin. In: Www.sciencedirect.com [online]. Amsterdam: Elsevier, 2014 [cit. 2019-04-27]. Available at: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/tafazzin </ref> Mutations in TAZ gene lead to tafazzin not working properly, immature cardiolipin accumulates whereas the level of cardiolipin is low (mature cardiolopin has four linoleic acid side chains).<ref name="cit2" /><ref name="cit3" /> Mitochondria in affected patients are not having a normal shape and functions. Reduced energy production of mitochondria results in apoptosis of cells in tissues with high energy demands, especially cardiac and skeletal muscles. Moreover abnormally shaped mitochondria in white blood cells may affect their ability to proliferate. This causes [https://en.wikipedia.org/wiki/Neutropenia neutropenia]- decreased amount of white blood cells leading to higher risk of infections. <ref name="cit9" /> | |
=== Symptoms === | === Symptoms === | ||
| - | *dilated cardiomyopathy (CMD) | + | *[https://en.wikipedia.org/wiki/Dilated_cardiomyopathy dilated cardiomyopathy (CMD)] |
| - | *endocardial fibroelastosis (EFE) | + | *[https://en.wikipedia.org/wiki/Endocardial_fibroelastosis endocardial fibroelastosis (EFE)] |
*a predominantly proximal skeletal myopathy | *a predominantly proximal skeletal myopathy | ||
*growth retardation | *growth retardation | ||
*neutropenia | *neutropenia | ||
| - | *organic aciduria | + | *[https://en.wikipedia.org/wiki/Organic_acidemia organic aciduria] |
| - | *excess of 3-methylglutaconic acid <ref name=" | + | *excess of 3-methylglutaconic acid <ref name="cit5" /> |
*characteristic facial features (tall and broad forehead, round face, full cheeks, prominent pointed chin, large ears, and deep-set eyes) <ref name="cit10" /> | *characteristic facial features (tall and broad forehead, round face, full cheeks, prominent pointed chin, large ears, and deep-set eyes) <ref name="cit10" /> | ||
| Line 78: | Line 74: | ||
=== Diagnosis === | === Diagnosis === | ||
| - | Urinary 3-methylglutaconic acid (3-MGC) can be increased 5- to 20-fold, urinary 3-methylglutaric acid and 2-ethylhydracrylic acid may be moderately increased. The diagnosis is established in patient with increased MLCL:CL ratio or detected TAZ pathogenic variant on molecular genetic testing. <ref name="cit10"> | + | Urinary 3-methylglutaconic acid (3-MGC) can be increased 5- to 20-fold, urinary 3-methylglutaric acid and 2-ethylhydracrylic acid may be moderately increased. The diagnosis is established in patient with increased MLCL:CL ratio or detected TAZ pathogenic variant on molecular genetic testing. <ref name="cit10">PMID: 25299040</ref> |
== 3D Structure: Homology Model == | == 3D Structure: Homology Model == | ||
| - | No empirical 3D structure for human tafazzin protein (UniProt Q16635) is available in April, 2019. In view of this, homology model was constructed using Swiss-Model server. <ref name="cit17">https://swissmodel.expasy.org/repository/uniprot/Q16635</ref> The closest related protein to tafazzin with a known 3D structure in a public database is in April 2019 plant glycerol 3-phosphate acyltransferase (G3PAT) which has sequence identity of 18,10 %, sequence similarity is 0,29. Tafazzin belongs to a protein superfamily - phospholipid acyltransferases (PF01553). | + | No empirical 3D structure for human tafazzin protein (UniProt Q16635) is available in April, 2019. In view of this, [https://proteopedia.org/wiki/index.php/Homology_modeling homology model] was constructed using Swiss-Model server. <ref name="cit17">Q16635 (TAZ_HUMAN) Homo sapiens (Human). In: Swissmodel.expasy.org/ [online]. Lausanne, Basel: swissmodel, 2019 [cit. 2019-04-27]. Available at: https://swissmodel.expasy.org/repository/uniprot/Q16635</ref> The closest related protein to tafazzin with a known 3D structure in a public database is in April 2019 plant glycerol 3-phosphate acyltransferase ([https://www.rcsb.org/structure/1iuq G3PAT]) which has sequence identity of 18,10 %, sequence similarity is 0,29. <ref name="cit17" /> Tafazzin belongs to a protein superfamily - phospholipid acyltransferases (PF01553).<ref name="cit25">Family: Acyltransferase (PF01553). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Available at: https://pfam.xfam.org/family/PF01553#tabview=tab0</ref> |
| - | Domains found on sequence of human tafazzin contain transmembrane domain (15-34) and acyltransferase domain (41-215). <ref name=" | + | Domains found on sequence of human tafazzin contain transmembrane domain (15-34) and acyltransferase domain (41-215). <ref name="cit19">Protein: TAZ_HUMAN (Q16635). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Available at: http://pfam.xfam.org/protein/Q16635</ref> |
| + | |||
[[Image:ConSurfTafazzin.png|thumb|Output from ConSurf analysis of tafazzin showing HX4D motif conservation.]] | [[Image:ConSurfTafazzin.png|thumb|Output from ConSurf analysis of tafazzin showing HX4D motif conservation.]] | ||
| - | [[Image:HMMLogoAcyltransferase.png|thumb|HMM logo of acyltransferase family (PF01553). <ref name="cit18">https://pfam.xfam.org/family/PF01553#tabview=tab4</ref>]] | + | [[Image:HMMLogoAcyltransferase.png|thumb|HMM logo of acyltransferase family (PF01553). <ref name="cit18">Family: Acyltransferase (PF01553). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Available at: https://pfam.xfam.org/family/PF01553#tabview=tab4</ref>]] |
| + | |||
| + | In the acyltransferase domain there is a HX<sub>4</sub>D [https://proteopedia.org/wiki/index.php/Introduction_to_protein_structure#Motifs_In_Proteins motif] - <scene name='81/813423/Tafazzinpredictedcartoonhis69/1'>histidine</scene> and <scene name='81/813423/Tafazzinpredictedcartoonasp74/1'>aspartic acid</scene> residues separated by four less conserved residues. Sequence analysis of membrane-bound glycerolipid acyltransferases revealed that proteins from the bacterial, plant, and animal kingdoms share this highly conserved domain containing <scene name='81/813423/Tafazzinpredictedcartoonhx4d/1'>HX<sub>4</sub>D motif</scene>. | ||
| + | <ref name="cit21">PMID: 9515909</ref> | ||
| - | + | Hijikatu, Yura, Ohara and Go in their work from 2015 predicted the [https://en.wikipedia.org/wiki/Intrinsically_disordered_proteins intrinsically unstructured regions] present in human tafazzin using 15 available prediction servers, eleven of them consistently predicted that the region encoded by exon 5 is intrinsically unstructured, and no other regions of the tafazzin sequence were consistently predicted as intrinsically unstructured.<ref name="cit8" /> | |
| - | + | ||
| - | Hijikatu, Yura, Ohara and Go in their work from 2015 predicted the intrinsically unstructured regions present in human tafazzin using 15 available prediction servers, eleven of them consistently predicted that the region encoded by exon 5 is intrinsically unstructured, and no other regions of the tafazzin sequence were consistently predicted as intrinsically unstructured. | + | |
| + | Hijikatu, Yura, Ohara and Go state in their work <scene name='81/813423/Tafazzinpredictedmutations/1'>33 known mutations</scene> of which 2 (Arg94 and Gly197) are replaced more than once. They divided mutated amino acids into 4 groups - amino acids associated with membranes (Asn40, Ile54, Arg57, Arg94, Lys117 - red colored), destabilization (Thr43, Leu50, Gly80, Leu82, Ile209, Leu210, Leu212, His214, Gly216, Gly240 - green colored), substrate binding (Pro62, Ser71, Asp101, Phe104, Ser110, Gly116, Cys118, Val119, Gly124, Phe108, Gly161, Leu169, Trp174, Phe178, Val183, Gly195, Gly197 - blue colored) and catalytic activity (His69 - yellow). <ref name="cit8" /> | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
This page is a project made with my classmates Kateřina Krausová and Dominik Merta for Structural Biology of the Cell Class in summer 2019 at the Charles University.
Tafazzin
| Theoretical Model: The protein structure described on this page was determined theoretically, and hence should be interpreted with caution. |
Tafazzin is a protein located in mitochondrial inner membranes. It is involved in altering cardiolipin. Cardiolipin is key in maintaining mitochondrial shape, energy production, and protein transport within cells. The full-length tafazzin protein contains 292 amino acids and has a molecular weight of 33459 daltons. Mutations in gene associated with this protein can cause Barth Syndrome. Barth syndrome (BTHS) is a genetic disorder diagnosed almost exclusively in males. BTHS is rare, it is estimated to affect 1 in 300,000 to 400,000 individuals worldwide. Males with BTHS have weak heart and skeletal muscles which can lead to heart failure. Another of the symptoms is neutropenia which can lead to infections. [1]
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Barth syndrome. In: Ghr.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2019 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/condition/barth-syndrome#
- ↑ 2.0 2.1 2.2 2.3 Q16635 (TAZ_HUMAN). In: Https://www.uniprot.org/ [online]. Cambridge, Geneva, Washington: UniProt, 2019 [cit. 2019-04-27]. Available at: https://www.uniprot.org/uniprot/Q16635?fbclid=IwAR3v10lUTRZfb0NFOYKC4wjaherdU9PIVJ8T63jkC9RfNu_5OQ2IpoDR0iY
- ↑ 3.0 3.1 3.2 3.3 CHRISTIE, William. Cardiolipin (Diphosphatidylglycerol). In: Www.lipidhome.co.uk [online]. Hutton: The LipidWeb, 2019 [cit. 2019-04-27]. Available at: http://www.lipidhome.co.uk/lipids/complex/dpg/index.htm
- ↑ 4.0 4.1 4.2 4.3 Houtkooper, R. H., & Vaz, F. M. (2008). Cardiolipin, the heart of mitochondrial metabolism. Cellular and Molecular Life Sciences, 65(16), 2493–2506. doi:10.1007/s00018-008-8030-5
- ↑ Cardiolipin structure. In: Commons.wikimedia.org [online]. San Francisco: Wikimedia, 2019 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Cardiolipin_structure.svg?uselang=cs
- ↑ Rochellehx. File:Eukaryotic pathway.jpg. In: Wikimedia Commons [online]. 23. 4. 2009 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Eukaryotic_pathway.jpg
- ↑ 7.0 7.1 7.2 Raja V, Greenberg ML. The functions of cardiolipin in cellular metabolism-potential modifiers of the Barth syndrome phenotype. Chem Phys Lipids. 2014 Apr;179:49-56. doi: 10.1016/j.chemphyslip.2013.12.009., Epub 2014 Jan 17. PMID:24445246 doi:http://dx.doi.org/10.1016/j.chemphyslip.2013.12.009
- ↑ TAZ gene. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2014 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/gene/TAZ#
- ↑ 9.0 9.1 9.2 Hijikata A, Yura K, Ohara O, Go M. Structural and functional analyses of Barth syndrome-causing mutations and alternative splicing in the tafazzin acyltransferase domain. Meta Gene. 2015 Apr 22;4:92-106. doi: 10.1016/j.mgene.2015.04.001. eCollection, 2015 Jun. PMID:25941633 doi:http://dx.doi.org/10.1016/j.mgene.2015.04.001
- ↑ 10.0 10.1 Barth syndrome cells display widespread remodeling of mitochondrial complexes without affecting metabolic flux distribution. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease [online]. 2018, 1864(11), 3650-3658 [cit. 2019-04-27]. Available at: https://www.sciencedirect.com/science/article/pii/S092544391830334X?via%3Dihub
- ↑ 11.0 11.1 Barth syndrome: an X‐linked cause of fetal cardiomyopathy and stillbirth. Prenatal Diagnosis [online]. 2010, 30(10), 970-976 [cit. 2019-04-27]. Available at: https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.2599
- ↑ Tafazzin. In: Www.sciencedirect.com [online]. Amsterdam: Elsevier, 2014 [cit. 2019-04-27]. Available at: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/tafazzin
- ↑ 13.0 13.1 13.2 Ferreira C, Thompson R, Vernon H. Barth Syndrome PMID:25299040
- ↑ 14.0 14.1 Q16635 (TAZ_HUMAN) Homo sapiens (Human). In: Swissmodel.expasy.org/ [online]. Lausanne, Basel: swissmodel, 2019 [cit. 2019-04-27]. Available at: https://swissmodel.expasy.org/repository/uniprot/Q16635
- ↑ Family: Acyltransferase (PF01553). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Available at: https://pfam.xfam.org/family/PF01553#tabview=tab0
- ↑ Protein: TAZ_HUMAN (Q16635). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Available at: http://pfam.xfam.org/protein/Q16635
- ↑ Family: Acyltransferase (PF01553). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Available at: https://pfam.xfam.org/family/PF01553#tabview=tab4
- ↑ Heath RJ, Rock CO. A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol. 1998 Mar;180(6):1425-30. PMID:9515909