Journal:CHEMBIOINT:1
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection load='' size='450' side='right' scene='81/817522/Cv/16' caption=''> | <StructureSection load='' size='450' side='right' scene='81/817522/Cv/16' caption=''> | ||
===The four-helix bundle in cholinesterase dimers: structural and energetic determinants of stability=== | ===The four-helix bundle in cholinesterase dimers: structural and energetic determinants of stability=== | ||
| - | <big>Dana A. Novichkova, Sofya V. Lushchekina, Orly Dym, Patrick Masson, Israel Silman and Joel L. Sussman</big> <ref> | + | <big>Dana A. Novichkova, Sofya V. Lushchekina, Orly Dym, Patrick Masson, Israel Silman and Joel L. Sussman</big> <ref>doi 10.1016/j.cbi.2019.06.012</ref> |
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| Line 13: | Line 13: | ||
All crystal structures of AChE deposited in the PDB (https://www.rcsb.org) display dimers in which the monomers associate through an antiparallel four-helix bundle. With respect to BChE, crystal structures have been obtained only for monomeric forms of hBChE. In the first hBChE crystal structure obtained (PDB ID [[1p0i]]), of partially glycosylated enzyme expressed in CHO cells, dimers assembling via a <scene name='81/817522/Cv/6'>four-helix bundle were not observed</scene>. Subsequently, however, a <scene name='81/817522/Cv/5'>crystal structure in which the dimers did assemble via a four-helix bundle</scene> (PDB ID [[4aqd]]) was obtained for fully glycosylated hBChE expressed in ''Drosophila S2'' cells. This dissimilarity of the two crystal structures reveals the high sensitivity of hBChE dimerization to the expression system and crystallization conditions. | All crystal structures of AChE deposited in the PDB (https://www.rcsb.org) display dimers in which the monomers associate through an antiparallel four-helix bundle. With respect to BChE, crystal structures have been obtained only for monomeric forms of hBChE. In the first hBChE crystal structure obtained (PDB ID [[1p0i]]), of partially glycosylated enzyme expressed in CHO cells, dimers assembling via a <scene name='81/817522/Cv/6'>four-helix bundle were not observed</scene>. Subsequently, however, a <scene name='81/817522/Cv/5'>crystal structure in which the dimers did assemble via a four-helix bundle</scene> (PDB ID [[4aqd]]) was obtained for fully glycosylated hBChE expressed in ''Drosophila S2'' cells. This dissimilarity of the two crystal structures reveals the high sensitivity of hBChE dimerization to the expression system and crystallization conditions. | ||
| - | ''In silico'' alanine screening (FEP method) for hAChE. <scene name='81/817522/Cv/10'>Top view of the dimerization interface of dimer</scene>. The amino acids, which substitution by alanine led |ΔΔG|>1 kcal/mol are shown and colored in green or red. <scene name='81/817522/Cv/ | + | ''In silico'' alanine screening (FEP method) for hAChE. <scene name='81/817522/Cv/10'>Top view of the dimerization interface of dimer</scene>. The amino acids, which substitution by alanine led |ΔΔG|>1 kcal/mol are shown and colored in green or red. <scene name='81/817522/Cv/19'>Side view of the dimerization interface of one of the monomers</scene>. These scenes show that in the case of hAChE, stabilization occurs with both hydrophobic residues of helices, and the salt bridge, and electrostatic interactions on helices. Mutations of His381 and Phe531 to alanine are advantageous. <scene name='81/817522/Cv/12'>These two residues are located in close proximity to each other</scene>, and their interactions appear to be stabilizing. |
| - | ''In silico'' alanine screening (FEP method) for BChE. <scene name='81/817522/Cv/15'>Top view of the dimerization interface of dimer</scene>. The amino acids, which substitution by alanine led |ΔΔG|>1 kcal/mol are shown and colored in green or red. <scene name='81/817522/Cv/ | + | ''In silico'' alanine screening (FEP method) for BChE. <scene name='81/817522/Cv/15'>Top view of the dimerization interface of dimer</scene>. The amino acids, which substitution by alanine led |ΔΔG|>1 kcal/mol are shown and colored in green or red. <scene name='81/817522/Cv/21'>Side view of the dimerization interface of one of the monomers</scene>. BChE has less stabilizing amino acids in loops compared to hAChE. Amino acids unfavorable for BChE dimerization are 1) polar ones (i.e., serine) instead of hydrophobic in helices 2) not exposed to the contact interface (Leu370) 3) Asp375 and Asp378, which repel each other and destabilize the loop. His372 in BChE (analog of His381 in hAChE) forms hydrogen bond with Gln518, and is more buried from the dimerization interface, thus it does not have the destabilizing effect observed in hAChE. Generally, ''in silico'' alanine screening shows that hAChE has less amino acid residues with high contribution to both stabilization and destabilization of the dimer, rather than BChE. In spite of a few residues on helices of BChE, extrahelical loops contribute to destabilization of the dimer. |
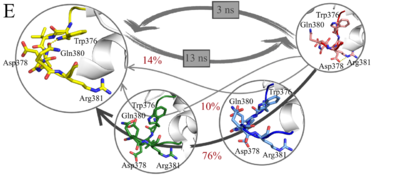
<scene name='81/817522/Cv1/2'>Macrostates. BChE dimer, top view</scene>. Red loop — first macrostate, blue loop — second macrostate, green loop — third macrostate, yellow loop - forth macrostate. <scene name='81/817522/Cv1/3'>Macrostates. BChE dimer, side view</scene>. | <scene name='81/817522/Cv1/2'>Macrostates. BChE dimer, top view</scene>. Red loop — first macrostate, blue loop — second macrostate, green loop — third macrostate, yellow loop - forth macrostate. <scene name='81/817522/Cv1/3'>Macrostates. BChE dimer, side view</scene>. | ||
| Line 28: | Line 28: | ||
This study was supported by Russian Foundation for Basic Research (project №19-03-00043). | This study was supported by Russian Foundation for Basic Research (project №19-03-00043). | ||
| + | |||
<b>References</b><br> | <b>References</b><br> | ||
<references/> | <references/> | ||
</StructureSection> | </StructureSection> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.