Dioxygenase
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection size="" scene ="Journal:JBIC:5/Opening/2" caption="Solved Crystal Structure of Hyperactive Catechol Dioxygenase"> | <StructureSection size="" scene ="Journal:JBIC:5/Opening/2" caption="Solved Crystal Structure of Hyperactive Catechol Dioxygenase"> | ||
| - | + | __TOC__ | |
== Function == | == Function == | ||
| Line 12: | Line 12: | ||
* '''Glyoxalase/bleomycin resistant protein/dioxygenase''' (GBD) catalyzes the cleavage of S-lactoyl-glutathione to methylglyoxal and glutathione.<br /> | * '''Glyoxalase/bleomycin resistant protein/dioxygenase''' (GBD) catalyzes the cleavage of S-lactoyl-glutathione to methylglyoxal and glutathione.<br /> | ||
* '''Methylcytosine dioxygenase''' (TET) catalyzes the conversion of 5-methylcytosine to 5-hydroxymethylcytosine to mediate DNA demethylation<ref>PMID:25862091</ref>. For details see [[TET Enzymes]].<br /> | * '''Methylcytosine dioxygenase''' (TET) catalyzes the conversion of 5-methylcytosine to 5-hydroxymethylcytosine to mediate DNA demethylation<ref>PMID:25862091</ref>. For details see [[TET Enzymes]].<br /> | ||
| + | * '''α-ketoglutarate-dependent dioxygenase Alkb''' repairs alkylated DNA and RNA containing 3-methylcytosine or 1-methyladenine by oxidative demethylation<ref>PMID:18432238</ref>.<br /> | ||
| + | * '''α-ketoglutarate-dependent dioxygenase Fto''' highest activity is repairing alkylated RNA containing 3-methyluracil by oxidative demethylation<ref>PMID:18775698</ref>.<br /> | ||
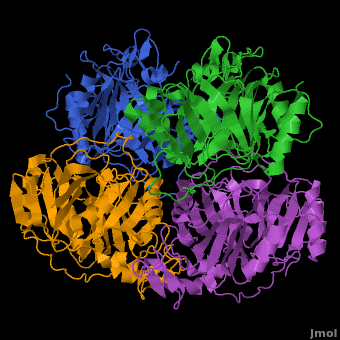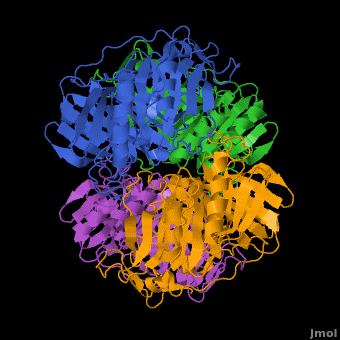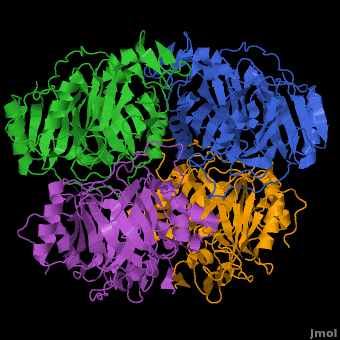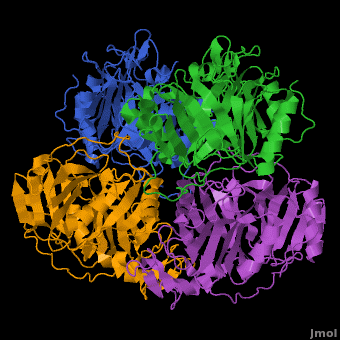
== A Hyperactive Cobalt-Substituted Extradiol-Cleaving Catechol Dioxygenase <ref>DOI 10.1007/s00775-010-0732-0</ref>== | == A Hyperactive Cobalt-Substituted Extradiol-Cleaving Catechol Dioxygenase <ref>DOI 10.1007/s00775-010-0732-0</ref>== | ||
| Line 23: | Line 25: | ||
</StructureSection> | </StructureSection> | ||
| - | + | ==References== | |
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, Tian Q, Aifantis I, Wei L, Dong C. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015 Apr 21;42(4):613-26. doi: 10.1016/j.immuni.2015.03.005. Epub 2015 , Apr 7. PMID:25862091 doi:http://dx.doi.org/10.1016/j.immuni.2015.03.005
- ↑ Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008 Apr 24;452(7190):961-5. PMID:18432238 doi:http://dx.doi.org/10.1038/nature06889
- ↑ Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008 Oct 15;582(23-24):3313-9. doi: 10.1016/j.febslet.2008.08.019., Epub 2008 Sep 5. PMID:18775698 doi:10.1016/j.febslet.2008.08.019
- ↑ Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L Jr. A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase. J Biol Inorg Chem. 2010 Dec 14. PMID:21153851 doi:10.1007/s00775-010-0732-0
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky