|
|
| (12 intermediate revisions not shown.) |
| Line 2: |
Line 2: |
| | | | |
| | == Function == | | == Function == |
| - | [[Heat Shock Proteins]] (Hsp). The Hsp are proteins which are expressed when cells are exposed to stresses like heat. Hsp60, Hsp70, Hsp90 etc. are named according to their molecular weights. Their function is to provide protection from stress<ref>PMID:18432918</ref>. Hsp contains a nucleotide-binding domain (NBD), a substrate-binding domain (SBD) and C-terminal lid domain (lid). In ''E. coli'' there are additional Hsp – HslV (19kD), HslJ and HslU (50kD). The heat-inducible Hsp is called '''heat shock cognate protein''' (Hsc). '''Hsp70''' from ''E. coli'' is called '''DnaK'''. '''Hsp90B1''' is also called '''endoplasmin''' and '''grp94'''. | + | [[Heat Shock Proteins]] (Hsp). The Hsp are proteins which are expressed when cells are exposed to stresses like heat. Hsp60, Hsp70, Hsp90 etc. are named according to their molecular weights. Their function is to provide protection from stress<ref>PMID:18432918</ref>. Hsp contains a nucleotide-binding domain (NBD), a substrate-binding domain (SBD) and C-terminal lid domain (lid). In ''E. coli'' there are additional Hsp – HslV (19kD), HslJ and HslU (50kD). |
| | + | * '''Heat shock cognate protein''' (Hsc) is heat-inducible. |
| | + | *'''Hsp70''' from ''E. coli'' is called '''DnaK'''. See also [[Molecular Playground/DnaK]] and [[Hsp70]]. |
| | + | * '''Hsp70 family protein 5''' or '''endoplasmic reticulum chaperone BIP''' is located in the lumen of the ER and its synthesis is markedly induced under conditions that lead to the accumulation of unfolded polypeptides in the ER<ref>apmid;10597629</ref> |
| | + | * '''Hsp90B1''' is also called '''endoplasmin''' and '''grp94'''. |
| | + | *'''DegP''' is a dual-function chaperon and protease. It degrades peripasmic proteins under stress conditions<ref>PMID:31738379</ref>. |
| | + | *'''heat shock cognate protein''' expression is not induced by heat stress but are rather constitutively induced. |
| | See also<br /> | | See also<br /> |
| | * [[Chaperones]]<br /> | | * [[Chaperones]]<br /> |
| | * [[Hsp100]]<br /> | | * [[Hsp100]]<br /> |
| | * [[Hsp40]] | | * [[Hsp40]] |
| - | * [[Molecular Playground/DnaK]] for Hsp70 in ''E. coli'' | |
| | * [[Molecular Playground/sHSP]] for small Hsp | | * [[Molecular Playground/sHSP]] for small Hsp |
| | + | * [[Molecular Playground/Hsp70-Hsp90]] |
| | | | |
| | == Conformational dynamics of full-length inducible human Hsp70 derived from microsecond molecular dynamics simulations in explicit solvent <ref>doi 10.1080/07391102.2012.726190</ref> == | | == Conformational dynamics of full-length inducible human Hsp70 derived from microsecond molecular dynamics simulations in explicit solvent <ref>doi 10.1080/07391102.2012.726190</ref> == |
| Line 36: |
Line 42: |
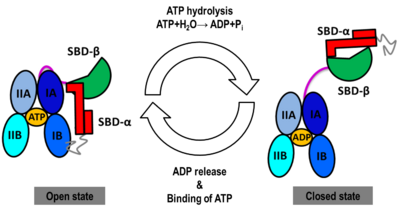
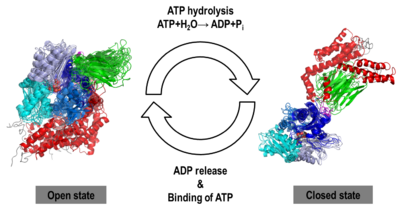
| | We found that human Hsp70, <scene name='Journal:JBSD:30/Cv/13'>both in its open and closed states</scene>, was better represented by an ensemble of conformations on a μs timescale, corresponding to different local minima of its free-energy landscape in good agreement with low-resolution experimental data of Hsp70s homologs (Small Angle Xray-Scattering and single-molecule Förster Resonance Energy Transfer) to which the MD simulations were compared in details. These structural ensembles represent the first attempt to model the conformational heterogeneity of a full-length molecular chaperone at the atomic scale. | | We found that human Hsp70, <scene name='Journal:JBSD:30/Cv/13'>both in its open and closed states</scene>, was better represented by an ensemble of conformations on a μs timescale, corresponding to different local minima of its free-energy landscape in good agreement with low-resolution experimental data of Hsp70s homologs (Small Angle Xray-Scattering and single-molecule Förster Resonance Energy Transfer) to which the MD simulations were compared in details. These structural ensembles represent the first attempt to model the conformational heterogeneity of a full-length molecular chaperone at the atomic scale. |
| | [[Image:JBSD30a.png|left|400px|thumb|<font color='blue'><b>NBD-IA is in blue</b></font>, <span style="color:deepskyblue;background-color:black;font-weight:bold;">NBD-IB is in deepskyblue</span>, <span style="color:lightsteelblue;background-color:black;font-weight:bold;">NBD-IIA is in lightsteelblue</span>, <span style="color:cyan;background-color:black;font-weight:bold;">NBD-IIB is in cyan</span>, <font color='magenta'><b>Linker is in magenta</b></font>, <span style="color:lime;background-color:black;font-weight:bold;">SBD-β is in green</span>, <font color='red'><b>SBD-α is in red</b></font>, <span style="color:darkgray;background-color:black;font-weight:bold;">C-terminal is in gray</span>]] | | [[Image:JBSD30a.png|left|400px|thumb|<font color='blue'><b>NBD-IA is in blue</b></font>, <span style="color:deepskyblue;background-color:black;font-weight:bold;">NBD-IB is in deepskyblue</span>, <span style="color:lightsteelblue;background-color:black;font-weight:bold;">NBD-IIA is in lightsteelblue</span>, <span style="color:cyan;background-color:black;font-weight:bold;">NBD-IIB is in cyan</span>, <font color='magenta'><b>Linker is in magenta</b></font>, <span style="color:lime;background-color:black;font-weight:bold;">SBD-β is in green</span>, <font color='red'><b>SBD-α is in red</b></font>, <span style="color:darkgray;background-color:black;font-weight:bold;">C-terminal is in gray</span>]] |
| - | </StructureSection>
| + | |
| | == 3D Structures of Heat Shock Proteins == | | == 3D Structures of Heat Shock Proteins == |
| | + | [[Heat Shock Protein structures]] |
| | | | |
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}}
| + | </StructureSection> |
| - | {{#tree:id=OrganizedByTopic|openlevels=0|
| + | |
| - | | + | |
| - | *Small Hsp
| + | |
| - | | + | |
| - | **[[1shs]], [[4eld]], [[4i88]] – Hsp – ''Methanococcus jannaschii''<br />
| + | |
| - | **[[1gme]] – wHsp 17kD – wheat<br />
| + | |
| - | **[[2byu]] - wHsp 17kD – Cryo EM<br />
| + | |
| - | **[[5ds1]] - pHsp 17.7 - pea<br />
| + | |
| - | **[[5ds2]] - pHsp 18.1<br />
| + | |
| - | **[[2bol]] – Hsp – ''Taenia saginata''<br />
| + | |
| - | **[[2h50]], [[2h53]] – yHsp26 – yeast<br />
| + | |
| - | **[[2l9q]], [[2ljl]], [[4axp]] – yHsp12 - NMR<br />
| + | |
| - | **[[2wj5]] – rHsp20 crystallin domain – rat<br />
| + | |
| - | **[[4rzk]], [[4yl9]] - SsHsp20 – ''Sulfolobus solfataricus''<br />
| + | |
| - | **[[4ylb]], [[4ylc]] - SsHsp20 (mutant)<br />
| + | |
| - | **[[2wj7]], [[3l1g]] – hHspB5 crystallin domain – human<br />
| + | |
| - | **[[4jus]] - hHsp20 6 residues 57-160<br />
| + | |
| - | **[[4jut]] - hHsp20 6 residues 57-160 (mutant)<br />
| + | |
| - | **[[5lum]] - hHsp20 6 ACD domain<br />
| + | |
| - | **[[5ltw]], [[5lu1]], [[5lu2]] - hHsp20 6 residues 1-149 + 14-3-3 sigma<br />
| + | |
| - | **[[3q9p]], [[3q9q]] – hHsp28 β1<br/>
| + | |
| - | **[[2n3j]], [[4mjh]] - hHsp28 1 crystallin domain<br />
| + | |
| - | **[[5z81]] – VcHsp15 – ''Vibrio cholerae''<br />
| + | |
| - | **[[3bbu]] – EcHsp15 - ''Escherichia coli'' – Cryo-EM<br />
| + | |
| - | **[[1ons]] – EcHsp31<br />
| + | |
| - | **[[3gla]], [[3gt6]], [[3guf]] – Hsp – ''Xanthomonas axonopodis''<br />
| + | |
| - | **[[3vqk]] – StHsp14 – ''Sulfolobus tokodaii''<br />
| + | |
| - | **[[3vql]], [[3vqm]] – StHsp14 (mutant)<br />
| + | |
| - | **[[3w1z]] – Hsp16 – ''Schizosacchromyces pombe''<br />
| + | |
| - | **[[5j7n]] – Hsp – ''Xylella fastidiosa''<br />
| + | |
| - | **[[5nms]] - Hsp 25.3 – ''Arabidopsis thaliana'' - Cryo EM<br />
| + | |
| - | | + | |
| - | *Hsp33
| + | |
| - | | + | |
| - | **[[1hw7]], [[3m7m]] – EcHsp33 NTD <br />
| + | |
| - | **[[1i7f]] – EcHsp33 chaperone domain (mutant)<br />
| + | |
| - | **[[1xjh]] - EcHsp33 CT - NMR<br />
| + | |
| - | **[[1vq0]] – Hsp33 – ''Thermotoga maritima''<br />
| + | |
| - | **[[1vzy]] – Hsp33 – ''Bacillus subtilis''
| + | |
| - | | + | |
| - | *HslV, HslU, HslJ
| + | |
| - | | + | |
| - | **[[1e94]], [[1yyf]], [[1g4a]], [[1g4b]], [[1hqy]], [[1ht1]], [[1ht2]], [[5ji3]] – EcHslV+HslU <br />
| + | |
| - | **[[5txv]] – EcHslU<br />
| + | |
| - | **[[5ji2]] – EcHslV + EcHslU (mutant)<br />
| + | |
| - | **[[1ned]] – EcHslV<br />
| + | |
| - | **[[4g4e]] – EcHslV (mutant)<br />
| + | |
| - | **[[2kts]] – EcHslJ - NMR<br />
| + | |
| - | **[[1g41]] – HiHslU – ''Haemophilus influenza''<br />
| + | |
| - | **[[1im2]] – HiHslU NBD
| + | |
| - | | + | |
| - | *Hsp40 or DnaJ homolog subfamily C member 12
| + | |
| - | | + | |
| - | **[[1c3g]] – yHsp40 CT<br />
| + | |
| - | **[[5vso]] – yHsp40 J domain – NMR<br />
| + | |
| - | **[[2qld]] – hHsp40<br />
| + | |
| - | **[[1hdj]] – hHsp40 J domain - NMR<br />
| + | |
| - | **[[1bq0]], [[1bqz]], [[1xbl]] – EcHsp40 N-terminal - NMR<br />
| + | |
| - | **[[1exk]] – EcHsp40 Cys-rich domain - NMR<br />
| + | |
| - | **[[4j7z]] – TtHsp40 residues 2-114 (mutant - ''Thermus thermophilus''<br />
| + | |
| - | | + | |
| - | *Hsp47
| + | |
| - | | + | |
| - | **[[3zha]] – dHsp47 + collagen peptide - dog<br />
| + | |
| - | | + | |
| - | * Hsp60 mitochondrial
| + | |
| - |
| + | |
| - | **[[4pj1]] – hHsp60 + hHsp10<br />
| + | |
| - | | + | |
| - | * Hsp60 - the large subunit of the chaperonin molecule GroEL
| + | |
| - | | + | |
| - | **[[3los]] – MmHsp60 - GroEL – ''Methanococcus maripaludis'' – closed state<br />
| + | |
| - | **[[3iyf]] - MmHsp60 - GroEL – open state<br />
| + | |
| - | **[[3e76]] – EcHsp60 – GroEL+Tl ion – ''Escherichia coli''<br />
| + | |
| - | **[[2nwc]], [[1oel]], [[1grl]], [[1la1]] - EcHsp60 – GroEL<br />
| + | |
| - | **[[2eu1]] - EcHsp60 (mutant) – GroEL<br />
| + | |
| - | **[[2cgt]] - EcHsp60 – GroEL+ADP+capsid assembly protein Gp31<br />
| + | |
| - | **[[2c7c]], [[2c7d]] - EcHsp60 – GroEL+GroES – cryo EM<br />
| + | |
| - | **[[4v4o]] - TtHsp60 – GroEL+GroES <br />
| + | |
| - | **[[1gr5]] - EcHsp60 – GroEL – cryo EM<br />
| + | |
| - | **[[1gru]] - EcHsp60 – GroEL+ATP+GroES+ADP – cryo EM<br />
| + | |
| - | **[[1dkd]] - EcHsp60 – GroEL apical (top) domain (146 amino acids)+12-mer peptide<br />
| + | |
| - | **[[1dk7]], [[1jon]] - EcHsp60 – GroEL apical domain <br />
| + | |
| - | **[[1srv]] - TtHsp60 – GroEL apical domain<br />
| + | |
| - | **[[1ass]], [[1asx]] - Hsp60 – GroEL apical domain – ''Thermoplasma acidophilum''<br />
| + | |
| - | **[[1kid]] - EcHsp60 (mutant) – GroEL apical domain<br />
| + | |
| - | | + | |
| - | * Hsp70 – 3 domains: N-terminus ATPase hydrolyzes ATP to ADP (NBD); hydrophobic amino acid binding domain (SBD) residues 381-564: C-terminus lid domain (CT) residues 534-641.
| + | |
| - | | + | |
| - | **[[5bn8]], [[2e88]], [[1s3x]], [[1hjo]] – hHsp70 NBD <br />
| + | |
| - | **[[5bn9]], [[5bpl]], [[5bpm]], [[5bpn]] – hHsp70 NBD (mutant)<br />
| + | |
| - | **[[3a8y]] – hHsp70 NBD+BAG domain regulator protein – human<br />
| + | |
| - | **[[4io8]], [[5aqw]], [[5aqx]], [[5aqy]], [[5aqz]], [[5ar0]], [[5mkr]], [[5mks]] - hHsp70 NBD + inhibitor<br />
| + | |
| - | **[[6b1i]] - hHsp70 NBD + ADP<br />
| + | |
| - | **[[6b1n]] - hHsp70 NBD (mutant) + ADP<br />
| + | |
| - | **[[6b1m]] - hHsp70 NBD (mutant) + ANP<br />
| + | |
| - | **[[2e8a]] - hHsp70 NBD+AMPPNP<br />
| + | |
| - | **[[3jxu]], [[3i33]], [[3gdq]], [[3fe1]] - hHsp70 NBD+ADP+P<br />
| + | |
| - | **[[3iuc]], [[3atu]], [[3atv]], [[3ay9]] - hHsp70 NBD+ADP<br />
| + | |
| - | **[[3lof]] – hHsp70 CT<br />
| + | |
| - | **[[4po2]] - hHsp70 CT + peptide<br />
| + | |
| - | **[[2lmg]] – hHsp70 CT - NMR<br />
| + | |
| - | **[[1xqs]] – hHsp70 SBD + Hsp-binding protein core domain<br />
| + | |
| - | **[[4j8f]], [[4wv5]], [[5xi9]], [[5gjj]] - hHsp70 SBD<br />
| + | |
| - | **[[5xir]] - hHsp70 SBD (mutant)<br />
| + | |
| - | **[[4wv7]] - hHsp70 SBD + novolactone<br />
| + | |
| - | **[[1hx1]] - bHsp70 NBD + molecular chaperone regulator 1 BAG domain<br />
| + | |
| - | **[[1q5l]] - EcHsp70 SBD +peptide - NMR<br />
| + | |
| - | **[[3gl1]] - ScHsp70 NBD – ''Saccharomyces cerevisiae''<br />
| + | |
| - | **[[3dob]], [[3dqg]], [[2op6]] - CeHsp70 SBD – ''Caenoharbditis elegans''<br />
| + | |
| - | **[[4i2w]] – CeHsp70 + myosin chaperone<br />
| + | |
| - | **[[2p32]] - CeHsp70 CT<br />
| + | |
| - | **[[2d2e]], [[3d2f]] – ScHsp70 NBD+SSE1(Hsp110)<br />
| + | |
| - | **[[2v7y]] – Hsp70 NBD+SBD – ''Geobacillus kaustophilus''<br />
| + | |
| - | **[[3l6q]] - CpHsp70 NBD – ''Cryptosporidium parvum''<br />
| + | |
| - | **[[3kvg]] - CpHsp70 NBD+AMPPNP<br />
| + | |
| - | **[[3l4i]] - CpHsp70 NBD+ADP+P<br />
| + | |
| - | | + | |
| - | *DnaK (Hsp70 from ''E. coli'') residues 389-607 SBD
| + | |
| - | | + | |
| - | **[[4b9q]] - EcDnaK NBD + SBD + ATP<br />
| + | |
| - | **[[5nro]] – EcDnaK (mutant) + DnaJ<br />
| + | |
| - | **[[2kho]] – EcDnaK – NMR<br />
| + | |
| - | **[[1dkg]] - EcDnaK NBD (mutant) + nucleotide exchange factor<br />
| + | |
| - | **[[4ani]] - EcDnaK NBD + SBD + nucleotide exchange factor<br />
| + | |
| - | **[[1dkx]], [[1dkz]] - EcDnaK NBD + sub<br />
| + | |
| - | **[[4jn4]], [[4jne]] - EcDnaK NBD + SBD (mutant) + ATP<br />
| + | |
| - | **[[4jnf]] - EcDnaK SBD (mutant) + ATP<br />
| + | |
| - | **[[1bpr]], [[1dg4]], [[2bpr]] - EcDnaK SBD – NMR<br />
| + | |
| - | **[[1q5l]] - EcDnaK SBD + peptide – NMR<br />
| + | |
| - | **[[3dpp]], [[3dpq]], [[3dpo]], [[1dky]], [[3qnj]], [[4e81]], [[4ezn]], [[4ezo]], [[4ezp]], [[4ezq]], [[4ezr]], [[4ezs]], [[4ezt]], [[4ezu]], [[4ezv]], [[4ezw]], [[4ezx]], [[4ezy]], [[4ezz]], [[4f00]], [[4hy9]], [[4hyb]], [[4jwc]], [[4jwd]], [[4jwe]], [[4jwi]], [[4r5i]], [[4f01]] – EcDnaK SBD + peptide<br />
| + | |
| - | **[[4r5j]], [[4r5k]], [[4r5l]] - EcDnaK CT <br />
| + | |
| - | **[[4r5g]] - EcDnaK CT + inhibitor<br />
| + | |
| - | **[[4rtf]] DnaK + ATP – ''Mycobacterium tuberculosis''<br />
| + | |
| - | **[[5obx]] - MgDnaK NBD – ''Mycoplasma genitalium''<br />
| + | |
| - | **[[5obv]], [[5obu]] - MgDnaK NBD (mutant) <br />
| + | |
| - | **[[5oby]] - MgDnaK NBD + AMPPNP <br />
| + | |
| - | **[[5obw]] - MgDnaK NBD + ATP <br />
| + | |
| - | | + | |
| - | *Hsp75
| + | |
| - |
| + | |
| - | **[[5f3k]], [[5f5r]] – hHsp75 NTD<br />
| + | |
| - | **[[5hph]] – hHsp75 residues 60-554<br />
| + | |
| - | **[[4z1f]], [[4z1g]], [[4z1h]], [[4z1i]], [[5y3n]], [[5y3o]] – hHsp75 + inhibitor<br />
| + | |
| - | | + | |
| - | *Hsp83
| + | |
| - | | + | |
| - | **[[3u67]] – LmHsp83 NTD + ADP – ''Leishmania major''
| + | |
| - | | + | |
| - | * Hsp90 – 4 domains: N-terminus (NTD) residues 1-236; linker; middle (MD); C-terminus (CT)
| + | |
| - | | + | |
| - | *''Hsp90-α NTD''
| + | |
| - | | + | |
| - | **[[3t0h]], [[5j2v]] - hHsp90 NTD – human<br />
| + | |
| - | **[[5j80]] - hHsp90 NTD (mutant)<br />
| + | |
| - | **[[2h8m]], [[2hch]], [[2gqp]] – dHsp90B1 NTD <br />
| + | |
| - | **[[1yt1]], [[1yt2]] - dHsp90B1 NTD (mutant) <br />
| + | |
| - | **[[3h80]] – LmHsp90 NTD <br />
| + | |
| - | **[[4gqt]] – CeHsp90 NTD<br />
| + | |
| - | **[[4x9l]] - Hsp90 NTD - rice<br />
| + | |
| - | | + | |
| - | *''Hsp90-α NTD complexes''
| + | |
| - | | + | |
| - | **[[3t0z]], [[3t2s]] - hHsp90-α NTD + ATP derivative<br />
| + | |
| - | **[[3t10]] - hHsp90-α NTD + ACP<br />
| + | |
| - | **[[3t1k]] - hHsp90-α NTD + ANP<br />
| + | |
| - | **[[3k97]], [[3hek]], [[3mnq]] – hHsp90 NTD+pyrrolidin derivative – human<br />
| + | |
| - | **[[3k98]], [[3k99]], [[3qtf]], [[5lo5]] - hHsp90 NTD+indole derivative<br />
| + | |
| - | **[[3r91]] - hHsp90 NTD (mutant) +indole derivative<br />
| + | |
| - | **[[3hhu]], [[4jql]] - hHsp90 NTD+ carbamic acid derivative<br />
| + | |
| - | **[[3ft8]], [[3ft5]], [[4cwf]], [[4cwn]], [[4cwo]], [[4cwp]], [[4cwq]], [[4cwr]], [[4cws]], [[4cwt]], [[6ei5]] - hHsp90 NTD+quinazoline derivative inhibitor<br />
| + | |
| - | **[[2k5b]] - hHsp90 NTD+Cdc37 – NMR - docking<br />
| + | |
| - | **[[3eko]], [[3ekr]] - hHsp90 NTD+dihydroxyphenyl amide<br />
| + | |
| - | **[[3bm9]], [[3bmy]] - hHsp90 NTD+benzisoxazole<br />
| + | |
| - | **[[2of6]], [[3b24]], [[3b27]], [[3b28]], [[3r92]] - hHsp90 NTD+triazine derivative<br />
| + | |
| - | **[[5j20]], [[5j27]], [[5j2x]], [[5j64]], [[5j6l]], [[5j6m]], [[5j82]], [[5j86]], [[5j8m]], [[5j8u]], [[5j9x]] - hHsp90-α NTD - hHsp90-a NTD+triazol derivative<br />
| + | |
| - | **[[2qfo]], [[3nmq]], [[3r4m]], [[3r4n]], [[3r4o]], [[3r4p]], [[3b25]], [[3b26]], [[2jcc]], [[3rlp]], [[3rlq]], [[3rlr]], [[2wi1]], [[2wi2]], [[2wi3]], [[2wi4]], [[2wi5]], [[2wi6]], [[2wi7]], [[2jjc]], [[5fnc]] - hHsp90 NTD+pyrimidine derivative<br />
| + | |
| - | **[[6f1n]] - hHsp90 NTD+pyridine derivative<br />
| + | |
| - | **[[1uy6]], [[1uy7]], [[1uy8]], [[1uy9]], [[1uyc]], [[1uyd]], [[1uye]], [[1uyf]], [[1uyg]], [[1uyh]], [[1uyi]], [[1uyk]], [[1uyl]], [[6el5]] - hHsp90 NTD+purine derivative<br />
| + | |
| - | **[[2qg0]], [[2bz5]] - hHsp90 NTD+sulfonamide derivative<br />
| + | |
| - | **[[2yi0]], [[2yi5]], [[2yi6]], [[2yi7]], [[5fnd]] - hHsp90 NTD + thiadiazole derivative<br />
| + | |
| - | **[[4hy6]], [[6ey8]], [[6ey9]], [[6eya]], [[6eyb]], [[5odx]], [[5oci]], [[5nyh]], [[5lny]], [[5lnz]], [[5lo0]], [[5lo1]], [[5lo6]] - hHsp90 NTD + indazole derivative<br />
| + | |
| - | **[[5nyi]] - hHsp90 NTD + resorcinol derivative<br />
| + | |
| - | **[[2qg2]], [[2cvi]], [[2cvj]], [[2uwd]], [[4b7p]] - hHsp90 NTD+oxazole derivative<br />
| + | |
| - | **[[3d0b]], [[3rkz]], [[3wq9]] - hHsp90 NTD+benzamide derivative<br />
| + | |
| - | **[[3inw]], [[3inx]] - hHsp90 NTD+pochoxime derivative<br />
| + | |
| - | **[[2fwy]], [[2fwz]], [[2h55]], [[3o0i]] - hHsp90 NTD+PU inhibitor<br />
| + | |
| - | **[[2ccs]], [[2cct]], [[2ccu]], [[2byh]], [[2byi]], [[2bsm]], [[2bt0]], [[1yc3]], [[1yc1]], [[1yc4]], [[6eln]], [[6elo]], [[6elp]] - hHsp90 NTD+pyrazol derivative<br />
| + | |
| - | **[[2ye2]], [[2ye3]], [[2ye4]], [[2ye5]], [[2ye6]], [[2ye7]], [[2ye8]], [[2ye9]], [[2yea]], [[2yeb]], [[2yec]], [[2yed]], [[2yee]], [[2yef]], [[2yeg]], [[2yeh]], [[2yei]], [[2yej]], [[2yjw]], [[2yjx]], [[2yk2]], [[2yk9]], [[2ykb]], [[2ykc]], [[2yke]], [[2yki]], [[2ykj]], [[3ow6]], [[3owb]], [[3owd]], [[3qdd]], [[3tuh]], [[3vha]], [[3vhc]], [[3vhd]], [[4awo]], [[4awp]], [[4awq]], [[4eeh]], [[4eft]], [[4efu]], [[4egh]], [[4egi]], [[4egk]], [[4fcp]], [[4fcq]], [[4fcr]], [[3wha]], [[4bqg]], [[4bqj]], [[4l8z]], [[4l90]], [[4l91]], [[4l93]], [[4lwe]], [[4l94]], [[4lwf]], [[4lwg]], [[4lwh]], [[4lwi]], [[4nh7]], [[4nh8]], [[4nh9]], [[4o09]], [[4o0b]], [[4r3m]], [[4u93]], [[4w7t]], [[4xip]], [[4xiq]], [[4xir]], [[4xit]], [[4ykq]], [[4ykr]], [[4ykt]], [[4yku]], [[4ykw]], [[4ykx]], [[4yky]], [[4ykz]], [[5cf0]], [[5fnf]], [[5ggz]], [[5m4e]], [[5m4h]], [[6ceo]], [[5vyy]], [[5t21]], [[5lq9]], [[5lr1]], [[5lr7]], [[5lrl]], [[5lrz]], [[5ls1]], [[5h22]] - hHsp90 NTD + inhibitor<br />
| + | |
| - | **[[1byq]] - hHsp90 NTD+ATP-Mg<br />
| + | |
| - | **[[2xhr]], [[2xht]], [[2xhx]], [[2xjg]], [[2xjj]], [[2xjx]], [[2xk2]], [[2xdx]], [[2xdu]], [[2xds]], [[2xdl]], [[2xdk]], [[2xab]], [[2vcj]], [[2vci]], [[3hyy]], [[3hyz]], [[3hz1]], [[3hz5]], [[3mnr]], [[2af6]] - hHsp90 NTD+ antigen<br />
| + | |
| - | **[[1osf]], [[1yer]], [[1yes]], [[1yet]] - hHsp90 NTD+geldamycin <br />
| + | |
| - | **[[3ied]] - PfHsp90 NTD+AMPPN – ''Plasmodium falciparum''<br />
| + | |
| - | **[[3k60]] - PfHsp90 NTD+ADP<br />
| + | |
| - | **[[2jki]] - Hsp90 NTD+SGT1 domain protein – barley<br />
| + | |
| - | **[[2hg1]], [[2gfd]], [[2fyp]], [[2exl]], [[1u0z]], [[1qy8]], [[1ysz]], [[1u2o]], [[6d28]], [[6d1x]], [[1qye]], [[3o2f]] - dHsp90B1 NTD+ligand<br />
| + | |
| - | **[[2esa]] - dHsp90B1 NTD (mutant) +ligand<br />
| + | |
| - | **[[1yt0]], [[1tc6]], [[1tba]], [[1tc0]] - dHsp90B1 NTD+ nucleotide<br />
| + | |
| - | **[[2xcm]] – Hsp90 NTD+CGT1+RAR1 – ''Hordum vulgare''<br />
| + | |
| - | **[[3q5j]], [[3q5k]], [[3q5l]] - LmHsp90 NTD + inhibitor<br />
| + | |
| - | **[[3o6o]], [[3omu]], [[3opd]] – TbHsp90 NTD+inhibitor – ''Trypanosoma brucei''<br />
| + | |
| - | **[[1a4h]] - yHsp90 NTD + antibiotic<br />
| + | |
| - | | + | |
| - | *''Hsp90-α MD''
| + | |
| - | | + | |
| - | **[[2gq0]] - EcHsp90 MD<br />
| + | |
| - | | + | |
| - | *''Hsp90-α CT''
| + | |
| - | | + | |
| - | **[[1sf8]] – EcHsp90 CT<br />
| + | |
| - | **[[3hjc]] - LmHsp90 CT<br />
| + | |
| - | | + | |
| - | *''Hsp90-α NTD + MD''
| + | |
| - | | + | |
| - | **[[2o1w]] - dHsp90B1 NTD+MD<br />
| + | |
| - | | + | |
| - | *''Hsp90-α MD + CT'' residues 293-732
| + | |
| - | | + | |
| - | **[[3q6m]], [[3q6n]] - hHsp90 MD + CT<br />
| + | |
| - | **[[2o1t]] – dHsp90B1 MD+CT<br />
| + | |
| - | | + | |
| - | *''Hsp90-α full length''
| + | |
| - | | + | |
| - | **[[1y4s]], [[1y4u]], [[2iop]], [[2ior]] - EcHsp90+ADP<br />
| + | |
| - | **[[2ioq]] - EcHsp90 <br />
| + | |
| - | **[[2o1u]] – dHsp90B1+AMPPNP<br />
| + | |
| - | **[[2o1v]] - dHsp90B1+ADP<br />
| + | |
| - | **[[2qf6]] - hHsp90 + antigen<br />
| + | |
| - | **[[5fwk]], [[5fwl]], [[5fwm]], [[5fwp]] – hHsp90 + CDC37 + CDK4<br />
| + | |
| - | | + | |
| - | *Hsp90-β
| + | |
| - | | + | |
| - | **[[5fwk]], [[5fwl]], [[5fwm]], [[5fwp]] – hHsp90 + CDC37 + CDK4<br />
| + | |
| - | **[[1uym]] - hHsp90 NTD+PU inhibitor<br />
| + | |
| - | **[[5uc4]], [[5uch]], [[5uci]], [[5ucj]], [[4o04]], [[4o05]], **[[4o07]] - hHsp90-b NTD + inhibitor<br />
| + | |
| - | **[[3nmq]] - hHsp90 NTD+pyrimidine derivative
| + | |
| - | **[[3pry]] – hHsp90 MD<br />
| + | |
| - | | + | |
| - | * Hsp82 is the yeast equivalent of Hsp90 in mammals
| + | |
| - | | + | |
| - | **[[1ah6]], [[1ah8]] – yHsp82 NTD<br />
| + | |
| - | **[[2xd6]], [[2xx2]], [[2xx4]], [[2xx5]], [[4ce1]], [[4ce2]], [[4ce3]] - yHsp82 NTD+macrolactone<br />
| + | |
| - | **[[1amw]] – yHsp82 NTD+ADP<br />
| + | |
| - | **[[2wep]] – yHsp82 NTD (mutant)+ADP<br />
| + | |
| - | **[[2akp]], [[3c0e]] - yHsp82 NTD (mutant)<br />
| + | |
| - | **[[1weq]], [[3c11]], [[2yga]], [[2wer]], [[2yge]], [[2ygf]] - yHsp82 NTD (mutant)+ antibiotic<br />
| + | |
| - | **[[2iws]], [[2iwu]], [[2iwx]], [[2cgf]], 1bgq, [[2fxs]], [[4as9]], [[4asa]], [[4asb]], [[4asf]], [[4asg]], [[2weq]], [[1ah4]], [[1bgq]] - yHsp82 NTD +antibiotic<br />
| + | |
| - | **[[2vw5]], [[2vwc]] - yHsp82 NTD +7-o-carbamoylpremacbecin<br />
| + | |
| - | **[[2fxs]] - yHsp82 NTD +radamide<br />
| + | |
| - | **[[1zw9]] - yHsp82 NTD + adenine derivative<br />
| + | |
| - | **[[1zwh]] - yHsp82 NTD + radester amine<br />
| + | |
| - | **[[2brc]], [[2bre]], [[1am1]] - yHsp82 NTD+pyrazol derivative<br />
| + | |
| - | **[[2cg9]] – yHsp82+co-chaperone SBA1<br />
| + | |
| - | **[[2cge]] - yHsp82 MD+CT+co-chaperone SBA1<br />
| + | |
| - | **[[1hk7]] - yHsp82 MD<br />
| + | |
| - | **[[1usu]], [[1usv]] - yHsp82 MD+co-chaperone AHA1<br />
| + | |
| - | **[[1us7]] - yHsp82 NTD+co-chaperone cdc37<br />
| + | |
| - | **[[3fp2]] – yHsp82 CT+TOM71
| + | |
| - | | + | |
| - | * Hsp100 are named ClpA or ClpB
| + | |
| - | | + | |
| - | **[[1r6b]], [[1ksf]] – EcClpA (mutant) <br />
| + | |
| - | **[[1r6c]], [[1k6k]] – EcClpA N-terminal <br />
| + | |
| - | **[[1r6o]], [[1r6q]] – EcClpA N-terminal + ClpS<br />
| + | |
| - | **[[1mg9]], [[1lzw]], [[1mbu]], [[1mbv]], [[1mbx]] – EcClpA + YLJA<br />
| + | |
| - | **[[4ciu]] - EcClpB (mutant)<br />
| + | |
| - | **[[4d2u]], [[4d2x]], [[4d2q]] - EcClpB – Cryo EM<br />
| + | |
| - | **[[1khy]] – EcClpB NTD <br />
| + | |
| - | **[[1jbk]] – EcClpB nucleotide binding domain 1<br />
| + | |
| - | **[[4iod]], [[4irf]] - PfClpB <br />
| + | |
| - | **[[1qvr]] - TtClpB <br />
| + | |
| - | **[[4fct]] - TtClpB CT<br />
| + | |
| - | **[[4lj4]] - TtClpB nucleotide binding domain 2<br />
| + | |
| - | **[[4hse]] - TtClpB nucleotide binding domain 1 + ADP<br />
| + | |
| - | **[[4lj5]] - TtClpB nucleotide binding domain 2 + ADP<br />
| + | |
| - | **[[4lj6]] - TtClpB nucleotide binding domain 2 + AMPPCP<br />
| + | |
| - | **[[4lj9]], [[4lja]] - TtClpB nucleotide binding domain 2 (mutant) + AMPPCP<br />
| + | |
| - | **[[4lj8]] - TtClpB nucleotide binding domain 2 (mutant) + ADP <br />
| + | |
| - | **[[4lj7]] - TtClpB nucleotide binding domain 2 (mutant) + ADP derivative<br />
| + | |
| - | **[[4fcv]], [[4fcw]], [[4fd2]] - TtClpB CT (mutant)<br />
| + | |
| - | **[[3zri]] - VcClpB NTD – ''Vibrio cholerae''<br />
| + | |
| - | **[[3zrj]] - VcClpB NTD + peptide<br />
| + | |
| - | | + | |
| - | *Hsp104
| + | |
| - | | + | |
| - | **[[5kne]] – yHsp104<br />
| + | |
| - | **[[5u2u]], [[6amn]], [[5wbw]] – yHsp104 NTD<br />
| + | |
| - | **[[5vy8]] – yHsp104 + ADP<br />
| + | |
| - | **[[5vy9]], [[5vya]] – yHsp104 + casein<br />
| + | |
| - | **[[5vjh]] – yHsp104 + casein + ATP-γ-S<br />
| + | |
| - | **[[5u2l]] – Hsp104 NTD – ''Candida albicans''<br />
| + | |
| - | | + | |
| - | * Hsp110 in mammals is named Sse1, Sse2 in yeast
| + | |
| - | | + | |
| - | **[[3d2e]], [[3d2f]], [[3c7n]] – ySse1+Hsc70 <br />
| + | |
| - | **[[2qxl]] – ySse1<br />
| + | |
| - | **[[2llv]] – ySti1 domain 1 – NMR<br />
| + | |
| - | **[[2llw]] – ySti1 domain 2 – NMR<br />
| + | |
| - | **[[3upv]] – ySti1 TPR repeats 7-9 + ySse1 peptide<br />
| + | |
| - | **[[3uq3]] - ySti1 TPR repeats 4-9 + peptide<br />
| + | |
| - | **[[3lca]] – ySsa1 lid +yTOM71<br />
| + | |
| - | | + | |
| - | *HspQ
| + | |
| - | | + | |
| - | **[[5ycq]] – EcHspQ<br />
| + | |
| - | | + | |
| - | *'''Heat shock cognate protein (Hsc)'''
| + | |
| - | | + | |
| - | *''Hsc70''
| + | |
| - | | + | |
| - | **[[3fzf]] - hHsc70 NBD+BAG domain regulator protein+ATP<br />
| + | |
| - | **[[3fzh]], [[3fzm]], [[3ldq]], [[3m3z]] - hHsc70 NBD+BAG domain regulator protein+ adenosine derivative<br />
| + | |
| - | **[[3fzk]], [[3fzl]] - hHsc70 NBD+BAG domain regulator protein+ purine derivative<br />
| + | |
| - | **[[3cqx]] - Hsc70 NBD+BAG domain regulator protein – mouse<br />
| + | |
| - | **[[4h5n]], [[4h5r]], [[4h5v]], [[4h5w]] - hHsc70 NBD<br />
| + | |
| - | **[[4h5t]] - hHsc70 NBD + ADP<br />
| + | |
| - | **[[4hwi]], [[5aqf]], [[5aqg]], [[5aqh]], [[5aqi]], [[5aqj]], [[5aqgk]], [[5aql]], [[5aqm]], [[5aqn]], [[5aqo]], [[5aqp]], [[5aqq]], [[5aqr]], [[5aqs]], [[5aqt]], [[5aqu]], [[5aqv]] - hHsc70 NBD+BAG domain regulator protein<br />
| + | |
| - | **[[2qw9]] – bHsc70 NBD – bovine<br />
| + | |
| - | **[[2bup]], [[1bup]], [[1qqm]], [[1qqn]], [[1qqo]] - bHsc70 NBD (mutant)<br />
| + | |
| - | **[[4fl9]], [[1yuw]] - bHsp70 NBD + SBD<br />
| + | |
| - | **[[2qwl]], [[2qwm]], [[1atr]], [[1ats]], [[1hpm]], [[1nga]], [[1ngb]], [[1ngc]], [[1ngd]], [[3hsc]] - bHsc70 NBD+ADP<br />
| + | |
| - | **[[2qwn]], [[2qwo]], [[2qwp]], [[2qwq]], [[2qwr]] - bHsc70 NBD (mutant)+ auxilin protein<br />
| + | |
| - | **[[1ba0]], [[1ba1]] – bHsc70 NBD (mutant) + ADP<br />
| + | |
| - | **[[1kax]], [[1kay]], [[1kaz]] - bHsc70 NBD (mutant) + ATP<br />
| + | |
| - | **[[1nge]], [[1ngf]], [[1ngg]], [[1ngh]] - bHsc70 NBD + ATP<br />
| + | |
| - | **[[1ngi]], [[1ngj]] - bHsc70 NBD + phosphoaminophosphonic acid<br />
| + | |
| - | **[[1hx1]] - bHsc70 NBD + molecular chaperone regulator 1 BAG domain<br />
| + | |
| - | **[[2v7z]] – rHsc70 NBD + SBD – rat<br />
| + | |
| - | **[[7hsc]], [[1ckr]] - rHsc70 sub - NMR<br />
| + | |
| - | | + | |
| - | *''Hsc90''
| + | |
| | | | |
| - | **[[4xkk]], [[4xe2]], [[4xka]], [[4xko]] – smHsc90 NTD – slime mold<br /> | |
| - | **[[4xcj]], [[4xc0]], [[4xcl]], [[4xd8]] – smHsc90 NTD + nucleotide<br /> | |
| - | **[[4xdm]] – smHsc90 NTD + antibiotic<br /> | |
| - | }} | |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | [[Category:Topic Page]] | | [[Category:Topic Page]] |
| | <references/> | | <references/> |