We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Cell division protein
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='2vam' size='340' side='right' caption='FtsZ of Bacillus subtilis' scene=''> | + | <StructureSection load='2vam' size='340' side='right' caption='FtsZ of Bacillus subtilis complex with sulfate (PDB code [[2vam]])' scene=''> |
== Bacillus subtilis division protein == | == Bacillus subtilis division protein == | ||
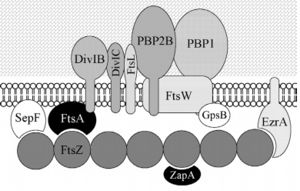
Bacillus subtilis is a prokaryotic organism, a Gram-positive bacterium, and thus has a cytoplasmic membrane plus a thick cell wall made of peptidoglycan and associated anionic polymers, such as teicoic acid. Bacteria are recognized for the reproductive success and conquest of various environments on Earth, with the presence of a complex and sophisticated machinery of cell division and formation of identical daughter cells a potent factor in this success. There are currently 24 proteins known to be associated with division in B. subtilis: ClpX, DivIB,DivIC, DivIVA, EzrA, FtsA, FtsL, FtsW, FtsZ,GpsB, MciZ, MinC, MinD, MinJ, Noc, PBP1,PBP2B, SepF, SftA, SpoIIE, SpoIIIE, UgtP, YneA and ZapA. These proteins can be divided in two main groups: proteins that make up the divisome - macromolecular complex composed of about 20 proteins, which promotes the construction of the cell wall and cytoplasmic membrane, forming the division septum (DivIB, DivIC, EzrA, FtsA, FtsL, FtsW,FtsZ, GpsB, PBP1, PBP2B, SepF and ZapA) and proteins that regulate the assembly of the divisome (ClpX, DivIVA, MciZ, MinC, MinD, MinJ,Noc, UgtP and YneA). | Bacillus subtilis is a prokaryotic organism, a Gram-positive bacterium, and thus has a cytoplasmic membrane plus a thick cell wall made of peptidoglycan and associated anionic polymers, such as teicoic acid. Bacteria are recognized for the reproductive success and conquest of various environments on Earth, with the presence of a complex and sophisticated machinery of cell division and formation of identical daughter cells a potent factor in this success. There are currently 24 proteins known to be associated with division in B. subtilis: ClpX, DivIB,DivIC, DivIVA, EzrA, FtsA, FtsL, FtsW, FtsZ,GpsB, MciZ, MinC, MinD, MinJ, Noc, PBP1,PBP2B, SepF, SftA, SpoIIE, SpoIIIE, UgtP, YneA and ZapA. These proteins can be divided in two main groups: proteins that make up the divisome - macromolecular complex composed of about 20 proteins, which promotes the construction of the cell wall and cytoplasmic membrane, forming the division septum (DivIB, DivIC, EzrA, FtsA, FtsL, FtsW,FtsZ, GpsB, PBP1, PBP2B, SepF and ZapA) and proteins that regulate the assembly of the divisome (ClpX, DivIVA, MciZ, MinC, MinD, MinJ,Noc, UgtP and YneA). | ||
| - | [[Image:DivisomeBacteria - Frederico.jpg | thumb | left | alt=Puzzle globe| B. subtilis divisome| 300 px]] Among these, the FtsZ stands out for acting in the recruitment of other proteins for the formation of the constriction ring that culminates in cell division. | + | [[Image:DivisomeBacteria - Frederico.jpg | thumb | left | alt=Puzzle globe| B. subtilis divisome| 300 px]] Among these, the '''FtsZ''' stands out for acting in the recruitment of other proteins for the formation of the constriction ring that culminates in cell division. |
| + | *'''FtsN''' is essential in activating septal peptidoglycan synthesis and constriction of the cell. | ||
| + | *'''FtsQ''' is essential in for early and late bacterial cell wall division<ref>PMID:18312270</ref>. | ||
| + | *'''FtsE''' and '''FtsX''' plays a central role by recruiting proteins to the divisive apparatus in bacteria <ref>PMID:32873757</ref> | ||
| + | *'''FtsY''' is a bacterial single-recognition particle receptor <ref>PMID:15815684</ref> | ||
| + | *'''ZapA''', '''ZapB''', '''ZapC''' and '''ZapD''' are recruited by FtsZ into the Z-ring forming during cell wall division<ref>PMID:20487275</ref>,<ref>PMID:22505682</ref>. | ||
| + | *'''ZipA''' links FtsZ to the cytoplasmic membrane<ref>PMID:10880432</ref>. | ||
| + | *'''SepF''' align FtsZ filaments in cyanobacteria<ref>PMID:24218584</ref>. | ||
| + | *'''DedA''' is part of the constriction phase of the bacterial cell division cycle<ref>PMID:30692172</ref>. | ||
== General Function == | == General Function == | ||
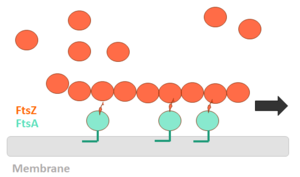
| - | FtsZ (Filamentation temperature sensitive Z) is the main coordinator of septum formation and the most widely conserved division protein, being present in essentially all bacterial genomes that have been sequenced to date. In eubacteria, the FtsZ gene is usually found in the dcw gene cluster, a DNA region containing division and cell-wall synthesis gene. This protein is extremely important to binary fission - more specifically, formation of the Z ring - in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Septum formation is faithfully coordinated with chromosome replication and segregation and the spatial control, on the other hand, is evident in the placement of the newly formed septum at precisely the middle of the progenitor cell, which ensures that the two daughter cells generated are morphologically and genetically equivalent. It means that despite the apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. | + | '''FtsZ''' (Filamentation temperature sensitive Z) is the main coordinator of septum formation and the most widely conserved division protein, being present in essentially all bacterial genomes that have been sequenced to date<ref>PMID:32692252</ref>. In eubacteria, the FtsZ gene is usually found in the dcw gene cluster, a DNA region containing division and cell-wall synthesis gene. This protein is extremely important to binary fission - more specifically, formation of the Z ring - in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Septum formation is faithfully coordinated with chromosome replication and segregation and the spatial control, on the other hand, is evident in the placement of the newly formed septum at precisely the middle of the progenitor cell, which ensures that the two daughter cells generated are morphologically and genetically equivalent. It means that despite the apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. |
== Structure and Biochemistry of FtsZ == | == Structure and Biochemistry of FtsZ == | ||
| - | FtsZ is a tubulin-like protein, which is widely conserved in bacteria and the main component of the bacterial cytokinesis machine, or “divisome.” FtsZ is a 40 kDa protein that folds into two independent globular domains [<scene name='81/817988/N-terminal_domain/1'>N-terminal</scene> (1-203) and <scene name='81/817988/C-terminal_domain/1'>C-terminal</scene> (204-316)] and has an unstructured tail of about 50 amino acids followed by a 15–17 conserved amino acid sequence at its extreme C-terminus. This conserved terminal sequence is also known as the ‘C-terminal peptide’ (CTP), since it is in the N-terminal domain that the nucleotide binding region is contained. Self-assembly of FtsZ involves interactions between the C-terminal globular domain of one subunit with the N-terminal globular domain of another subunit. The CTP, on the other hand, is the binding site for several of the proteins that interact with FtsZ. The N-terminal and C-terminal domain are separated by the central <scene name='81/817988/H7helix/1'>H7 helix</scene> (178-202). | + | '''FtsZ''' is a tubulin-like protein, which is widely conserved in bacteria and the main component of the bacterial cytokinesis machine, or “divisome.” FtsZ is a 40 kDa protein that folds into two independent globular domains [<scene name='81/817988/N-terminal_domain/1'>N-terminal</scene> (1-203) and <scene name='81/817988/C-terminal_domain/1'>C-terminal</scene> (204-316)] and has an unstructured tail of about 50 amino acids followed by a 15–17 conserved amino acid sequence at its extreme C-terminus. This conserved terminal sequence is also known as the ‘C-terminal peptide’ (CTP), since it is in the N-terminal domain that the nucleotide binding region is contained. Self-assembly of FtsZ involves interactions between the C-terminal globular domain of one subunit with the N-terminal globular domain of another subunit. The CTP, on the other hand, is the binding site for several of the proteins that interact with FtsZ. The N-terminal and C-terminal domain are separated by the central <scene name='81/817988/H7helix/1'>H7 helix</scene> (178-202). |
FtsZ and tubulin share several essential properties: their assembly is cooperative, stimulated by GTP, and leads to GTP hydrolysis; they form dynamic polymers whose turnover is dependent on nucleotide hydrolysis; they use essentially the same bond for polymer formation; and recent evidence indicates that they undergo similar allosteric transitions upon polymerization. The folding of the FtsZ N-terminal domain is typical of GTPases, with six parallel β-strands (S1-S6) surrounded by six α-helices (H1-H6), named according to the tubulin structure (show <scene name='81/817988/Secondarystructure/2'>secondary structure</scene>). The C-terminal domain is formed by four parallel β-strands (S7-S10) surrounded by two helices, with the antiparallel strand S10. The residues in the T1-T4 loops make contact with the phosphate groups of the GDP. The T5 loop between S5 and H5 helix contains residues that make hydrogen bonds with the sugar moiety and also contacts with the phosphate of GDP, while interactions with the nucleotide nitrogen base are done by residues of the H7 helix. | FtsZ and tubulin share several essential properties: their assembly is cooperative, stimulated by GTP, and leads to GTP hydrolysis; they form dynamic polymers whose turnover is dependent on nucleotide hydrolysis; they use essentially the same bond for polymer formation; and recent evidence indicates that they undergo similar allosteric transitions upon polymerization. The folding of the FtsZ N-terminal domain is typical of GTPases, with six parallel β-strands (S1-S6) surrounded by six α-helices (H1-H6), named according to the tubulin structure (show <scene name='81/817988/Secondarystructure/2'>secondary structure</scene>). The C-terminal domain is formed by four parallel β-strands (S7-S10) surrounded by two helices, with the antiparallel strand S10. The residues in the T1-T4 loops make contact with the phosphate groups of the GDP. The T5 loop between S5 and H5 helix contains residues that make hydrogen bonds with the sugar moiety and also contacts with the phosphate of GDP, while interactions with the nucleotide nitrogen base are done by residues of the H7 helix. | ||
| Line 19: | Line 27: | ||
== Relevance == | == Relevance == | ||
The understanding of the structure of FtsZ and its polymerization process is relevant because it can be used as a target for the design of new antibiotics capable of selectively combating bacterial infections. This is due to its structural and functional conservation in prokaryotes, to their essential role in the replication of these organisms, and the evolutionarily distant from tubulin, its counterpart in eukaryotes. Thus, the use of an antibiotic would not cause effects on eukaryotic tubulin. | The understanding of the structure of FtsZ and its polymerization process is relevant because it can be used as a target for the design of new antibiotics capable of selectively combating bacterial infections. This is due to its structural and functional conservation in prokaryotes, to their essential role in the replication of these organisms, and the evolutionarily distant from tubulin, its counterpart in eukaryotes. Thus, the use of an antibiotic would not cause effects on eukaryotic tubulin. | ||
| - | + | == 3D structures of cell division protein== | |
| + | [[Cell division protein 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
Current revision
| |||||||||||
References
[10] [11] [12] [13] [14] [15] [16] [17]
- ↑ van den Ent F, Vinkenvleugel TM, Ind A, West P, Veprintsev D, Nanninga N, den Blaauwen T, Lowe J. Structural and mutational analysis of the cell division protein FtsQ. Mol Microbiol. 2008 Apr;68(1):110-23. Epub 2008 Feb 26. PMID:18312270 doi:http://dx.doi.org/10.1111/j.1365-2958.2008.06141.x
- ↑ Alcorlo M, Straume D, Lutkenhaus J, Havarstein LS, Hermoso JA. Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae. mBio. 2020 Sep 1;11(5). pii: mBio.01488-20. doi: 10.1128/mBio.01488-20. PMID:32873757 doi:http://dx.doi.org/10.1128/mBio.01488-20
- ↑ Angelini S, Deitermann S, Koch HG. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 2005 May;6(5):476-81. PMID:15815684 doi:10.1038/sj.embor.7400385
- ↑ Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol. 2010 Jun;76(6):1514-26. PMID:20487275 doi:10.1111/j.1365-2958.2010.07183.x
- ↑ Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol. 2012 Jun;194(12):3189-98. doi: 10.1128/JB.00176-12. Epub 2012 Apr, 13. PMID:22505682 doi:http://dx.doi.org/10.1128/JB.00176-12
- ↑ Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000 Jul 3;19(13):3179-91. PMID:10880432 doi:10.1093/emboj/19.13.3179
- ↑ Duman R, Ishikawa S, Celik I, Strahl H, Ogasawara N, Troc P, Lowe J, Hamoen LW. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc Natl Acad Sci U S A. 2013 Nov 11. PMID:24218584 doi:http://dx.doi.org/10.1073/pnas.1313978110
- ↑ Liu B, Hale CA, Persons L, Phillips-Mason PJ, de Boer PAJ. Roles of the DedD Protein in Escherichia coli Cell Constriction. J Bacteriol. 2019 Mar 26;201(8):e00698-18. PMID:30692172 doi:10.1128/JB.00698-18
- ↑ Silber N, Matos de Opitz CL, Mayer C, Sass P. Cell division protein FtsZ: from structure and mechanism to antibiotic target. Future Microbiol. 2020 Jun;15:801-831. doi: 10.2217/fmb-2019-0348. Epub 2020 Jul , 21. PMID:32692252 doi:http://dx.doi.org/10.2217/fmb-2019-0348
- ↑ Bisson-Filho AW, Discola KF, Castellen P, Blasios V, Martins A, Sforca ML, Garcia W, Zeri AC, Erickson HP, Dessen A, Gueiros-Filho FJ. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci U S A. 2015 Apr 6. pii: 201414242. PMID:25848052 doi:http://dx.doi.org/10.1073/pnas.1414242112
- ↑ Wang, X. & Lutkenhaus, J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria.Mol. Microbiol. 21, 313–319 (1996). Gueiros-Filho, F. J. & Losick, R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002).
- ↑ Wang, X., Huang, J., Mukherjee, A., Cao, C., and Lutkenhaus, J. (1997). Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179, 5551–5559.
- ↑ Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004 Jan;58(1):19-29. doi: 10.1007/s00239-003-2523-5. PMID:14743312 doi:http://dx.doi.org/10.1007/s00239-003-2523-5
- ↑ Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014 Dec 9;3:e04601. doi: 10.7554/eLife.04601. PMID:25490152 doi:http://dx.doi.org/10.7554/eLife.04601
- ↑ Huecas, S. et al. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys. J. 94, 1796–1806 (2008).
- ↑ Lan, G. et al. (2009) Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 106, 121–126
- ↑ FILHO, Frederico Gueiros. Cell Division. In: GRAUMANN, Peter L. et al. Bacillus: Cellular and Molecular Biology. Germany: Caister Academic Press, 2017.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Jonathan Cardoso C. Vieira, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky