User:Luis Andres Casavilca Ramirez/Sandbox 1
From Proteopedia
< User:Luis Andres Casavilca Ramirez(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 9: | Line 9: | ||
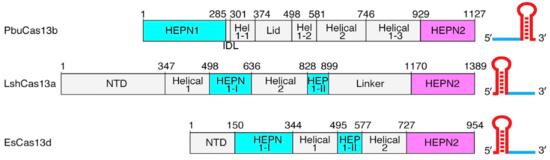
| - | Though the overall conserved bilobed shape of many Class 2 effectors consisting of a recognition and nuclease lobe is present,(2,3) Cas13b has a unique domain organization even among Cas13s. (Fig.1). A crystal structure of PbuCas13b complexed with a 36-nt direct repeat sequence and a 5-nt spacer at 1.65 Å provides information of the <scene name='81/817991/ | + | Though the overall conserved bilobed shape of many Class 2 effectors consisting of a recognition and nuclease lobe is present,(2,3) Cas13b has a unique domain organization even among Cas13s. (Fig.1). A crystal structure of PbuCas13b complexed with a 36-nt direct repeat sequence and a 5-nt spacer at 1.65 Å provides information of the <scene name='81/817991/Pbucas13b_domains3/1'>domains’ three dimensional arrange</scene>. There are five domains within the effector structure: two HEPN domains (<scene name='81/817991/Hepn1_and_hepn2/1'>HEPN1 and HEPN2</scene>), two mainly helical domains (<scene name='81/817991/Helical_1_and_helical_2/4'>Helical-1 and Helical-2</scene>), and a <scene name='81/817991/Lid_domain/2'>Lid domain</scene>. In addition, crRNA also has a unique structure, since its direct repeat is at the 3’ end and not at the 5’ end as in the other Cas13s (Fig.1). |
[[Image:Image Cas13 domains.png|550px|right|thumb| Fig.1 Linear domain organization of PbuCas13b, LshCas13a, and EsCas13d]] | [[Image:Image Cas13 domains.png|550px|right|thumb| Fig.1 Linear domain organization of PbuCas13b, LshCas13a, and EsCas13d]] | ||
| Line 35: | Line 35: | ||
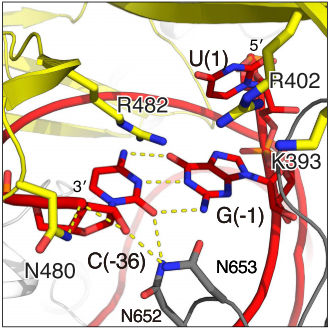
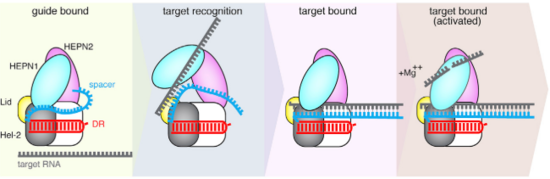
| - | The Cas13b-crRNA complex is practically inaccessible for hybridization with target RNA unless a major conformational shift allows access to the central channel. An opening between the domains HEPN1 and Helical-2 has been suggested, since it would provide a sterically permissible route with charged amino acids that would direct the RNA to the Cas13b central cavity. Mutation in a residue on a Lid domain β-hairpin (D397) at the interface between HEPN1 and Helical2 decreases Cas13 knockdown activity, suggesting an additional role of this domain in directing the target RNA into the central channel. | + | The <scene name='81/817991/Pbucas13b_domains_form_trans3/1'>Cas13b-crRNA complex</scene> is practically inaccessible for hybridization with target RNA unless a major conformational shift allows access to the central channel. An opening between the domains HEPN1 and Helical-2 has been suggested, since it would provide a sterically permissible route with charged amino acids that would direct the RNA to the Cas13b central cavity. Mutation in a residue on a Lid domain β-hairpin (D397) at the interface between HEPN1 and Helical2 decreases Cas13 knockdown activity, suggesting an additional role of this domain in directing the target RNA into the central channel. |
[[Image:Selection_039.png|550px|right|thumb| Fig.4 Proposed mechanism of crRNA targeting by Cas13b]] | [[Image:Selection_039.png|550px|right|thumb| Fig.4 Proposed mechanism of crRNA targeting by Cas13b]] | ||
| Line 47: | Line 47: | ||
== Cas13b engineering == | == Cas13b engineering == | ||
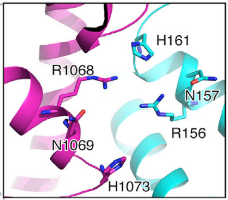
| - | [[Image:ActLoop.png| | + | [[Image:ActLoop.png|550px|left|thumb| Fig.5 HEPN2 conserved loop at active site]] |
The aforementioned preference for target RNA cleavage at dinucleotide sites is somewhat conserved among Cas 13 enzymes. A beta-hairpin loop located near the active site at HEPN2 domain is conserved among Cas13 from different species. Though the identities of its residues are variable, it is always located between highly conserved amino-acids (2) In PbuCas13b, mutants with altered residues at this loop that retained cleavage activity showed different cleavage preferences, and some of them even had increased preference for UU dinucleotides. SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unLOCKing), an mRNA detection technique that takes advantage of the promiscuous cleavage activity of Cas13s is limited by the nucleobases identities of some of transcripts.(8) Therefore, Cas13b engineering at this conserved loop could potentially increase the repertoire of substrates for these type of techniques. | The aforementioned preference for target RNA cleavage at dinucleotide sites is somewhat conserved among Cas 13 enzymes. A beta-hairpin loop located near the active site at HEPN2 domain is conserved among Cas13 from different species. Though the identities of its residues are variable, it is always located between highly conserved amino-acids (2) In PbuCas13b, mutants with altered residues at this loop that retained cleavage activity showed different cleavage preferences, and some of them even had increased preference for UU dinucleotides. SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unLOCKing), an mRNA detection technique that takes advantage of the promiscuous cleavage activity of Cas13s is limited by the nucleobases identities of some of transcripts.(8) Therefore, Cas13b engineering at this conserved loop could potentially increase the repertoire of substrates for these type of techniques. | ||
| Line 55: | Line 55: | ||
<references/> | <references/> | ||
<scene name='81/817991/Pbucas13b_central_channel/1'>Text To Be Displayed</scene> | <scene name='81/817991/Pbucas13b_central_channel/1'>Text To Be Displayed</scene> | ||
| + | <scene name='81/817991/Pbucas13b_domains_form/1'>Text To Be Displayed</scene> | ||
| + | <scene name='81/817991/Pbucas13b_domains3/1'>Text To Be Displayed</scene> | ||
| + | <scene name='81/817991/Pbucas13b_domains_form_trans3/1'>Text To Be Displayed</scene> | ||
Current revision
==Cas13b==
| |||||||||||
References