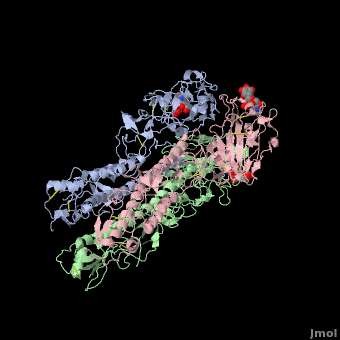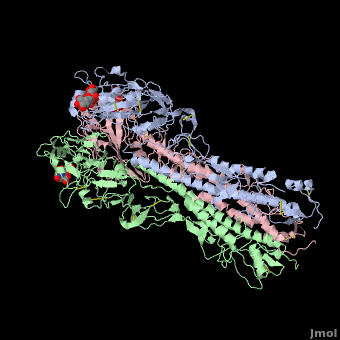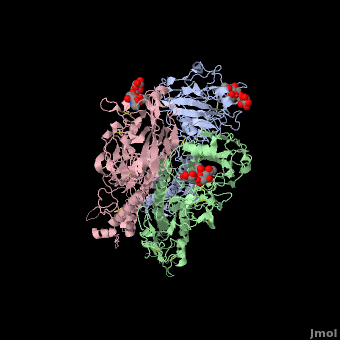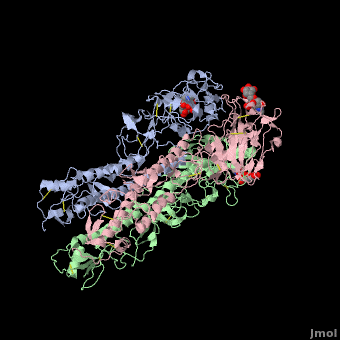
Hemagglutinin
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
1. On the target cell, HA binds the receptor on the cell membrane which is contains sialic acid <ref>Gottschalk A. Chemistry of virus receptors. The Viruses. 1959;3:51–61.</ref> allowing the virus/cell interaction. | 1. On the target cell, HA binds the receptor on the cell membrane which is contains sialic acid <ref>Gottschalk A. Chemistry of virus receptors. The Viruses. 1959;3:51–61.</ref> allowing the virus/cell interaction. | ||
2. HA induce the fusion between the host cell and the virus which can entry into the cytoplasm. | 2. HA induce the fusion between the host cell and the virus which can entry into the cytoplasm. | ||
| - | Because the HA are the major Antigens of the Virus, [[Antibody|Antibodies]] recognized them and for this reason HA often change. | + | Because the HA are the major Antigens of the Virus, [[Antibody|Antibodies]] recognized them and for this reason HA often change. |
| + | * '''Dr hemagglutinin''' is found in urinary tract pathogenic ''E. coli'' and recognizes Dr blood group antigen<ref>PMID: 3054548</ref>. | ||
| + | * '''Phytohemagglutinin''' is found in bean seeds and has sugar binding and hemagglutinin functions<ref>PMID: 8702788</ref>. | ||
| + | * '''Hemagglutinin-neuraminidase''' (HN) is multifunctional. It possesses both the receptor recognition and neuraminidase activities<ref>PMID:21680512</ref>. For details see [[Mumps Virus Hemagglutinin Neuraminidase Protein]]. | ||
For discussion of influenza hemagglutinin see<br /> | For discussion of influenza hemagglutinin see<br /> | ||
| Line 72: | Line 75: | ||
==3D structures of hemagglutinin== | ==3D structures of hemagglutinin== | ||
[[Hemagglutinin 3D structures]] | [[Hemagglutinin 3D structures]] | ||
| + | |||
| + | ==3D Printed Physical Models of Hemagglutinin== | ||
| + | |||
| + | Shown below are 3D printed physical models of Hemagglutinin. The model is shown as an alpha carbon backbone, with key sidechains and domains highlighted. It has been designed with precisely embedded magnets that allow the three chains to pull apart into individual pieces. | ||
| + | |||
| + | |||
| + | [[Image:hemagglutinin_1_centerForBiomolecularModeling.jpg | 450px]][[Image:hemagglutinin_2_centerForBiomolecularModeling.jpg | 450px]] | ||
| + | |||
| + | ====The MSOE Center for BioMolecular Modeling==== | ||
| + | |||
| + | [[Image:CbmUniversityLogo.jpg | left | 100px]] | ||
| + | |||
| + | The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. | ||
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Influenza
References
- ↑ Gottschalk A. Chemistry of virus receptors. The Viruses. 1959;3:51–61.
- ↑ Nowicki B, Hull R, Moulds J. Use of the Dr hemagglutinin of uropathogenic Escherichia coli to differentiate normal from abnormal red cells in paroxysmal nocturnal hemoglobinuria. N Engl J Med. 1988 Nov 10;319(19):1289-90. PMID:3054548 doi:10.1056/NEJM198811103191916
- ↑ Hamelryck TW, Dao-Thi MH, Poortmans F, Chrispeels MJ, Wyns L, Loris R. The crystallographic structure of phytohemagglutinin-L. J Biol Chem. 1996 Aug 23;271(34):20479-85. PMID:8702788
- ↑ Kim SH, Subbiah M, Samuel AS, Collins PL, Samal SK. Roles of the fusion and hemagglutinin-neuraminidase proteins in replication, tropism, and pathogenicity of avian paramyxoviruses. J Virol. 2011 Sep;85(17):8582-96. doi: 10.1128/JVI.00652-11. Epub 2011 Jun 15. PMID:21680512 doi:http://dx.doi.org/10.1128/JVI.00652-11
- ↑ Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365-94. PMID:3304138 doi:http://dx.doi.org/10.1146/annurev.bi.56.070187.002053
- ↑ Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17175-80. Epub 2009 Sep 28. PMID:19805083
- ↑ Cell Binding protein in Avian Influenza; Jack Cerchiara,'06 and Brendan Holsberry, 07;http://biology.kenyon.edu/BMB/Chime2/2005/Cerchiara-Holsberry/FRAMES/start.htm
- ↑ 8.0 8.1 Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006 Sep;119(1):1-7. PMID:16925526 doi:http://dx.doi.org/10.1111/j.1365-2567.2006.02421.x
- ↑ See Influenza in Wikipedia.
Proteopedia Page Contributors and Editors (what is this?)
Julien Madouasse, Michal Harel, David Canner, Mark Hoelzer, Ann Taylor, Alexander Berchansky, Joel L. Sussman