We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1n88
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==NMR structure of the ribosomal protein L23 from Thermus thermophilus.== | ==NMR structure of the ribosomal protein L23 from Thermus thermophilus.== | ||
| - | <StructureSection load='1n88' size='340' side='right'caption='[[1n88 | + | <StructureSection load='1n88' size='340' side='right'caption='[[1n88]]' scene=''> |
== Structural highlights == | == Structural highlights == | ||
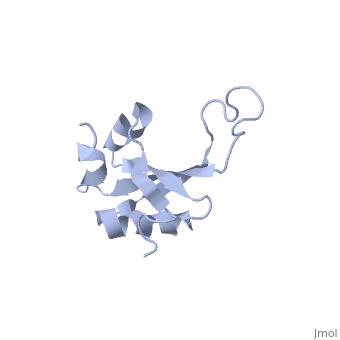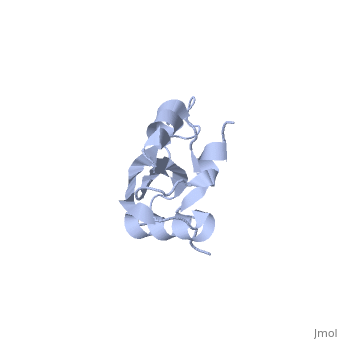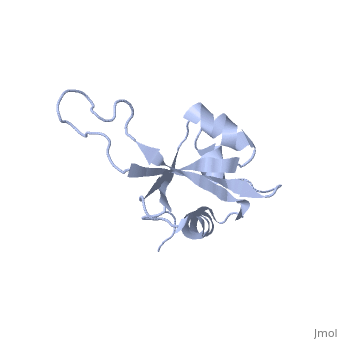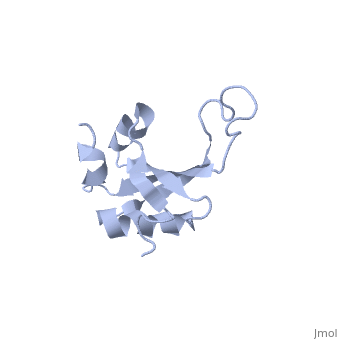
| - | <table><tr><td colspan='2'>[[1n88]] is a 1 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1n88]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Thermus_thermophilus Thermus thermophilus]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1N88 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1N88 FirstGlance]. <br> |
| - | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR</td></tr> |
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1n88 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1n88 OCA], [https://pdbe.org/1n88 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1n88 RCSB], [https://www.ebi.ac.uk/pdbsum/1n88 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1n88 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/RL23_THETH RL23_THETH] One of the early assembly proteins it binds 23S rRNA. One of the proteins that surrounds the polypeptide exit tunnel on the outside of the ribosome. Forms the main docking site for trigger factor binding to the ribosome (By similarity). |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 34: | Line 35: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: Flavobacterium thermophilum yoshida and oshima 1971]] | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: Dontsova | + | [[Category: Thermus thermophilus]] |
| - | [[Category: Garber | + | [[Category: Dontsova M]] |
| - | [[Category: Hard | + | [[Category: Garber MB]] |
| - | [[Category: Ohman | + | [[Category: Hard T]] |
| - | [[Category: Rak | + | [[Category: Ohman A]] |
| - | + | [[Category: Rak A]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
NMR structure of the ribosomal protein L23 from Thermus thermophilus.
| |||||||||||