Sandbox Reserved 1568
From Proteopedia
(Difference between revisions)
| (39 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_BHall_Chem351_F19}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_BHall_Chem351_F19}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | == | + | == '''Lignostilbene-α,ß-dioxygenase A structural features and important functional residues''' == |
| - | <StructureSection load=' | + | <StructureSection load='6ojt' size='420' side='right' caption='LsdA phenylazophenol complex' scene=''> |
| - | + | ||
| - | + | ||
| - | == | + | == '''Functions and Biological Relevance''' == |
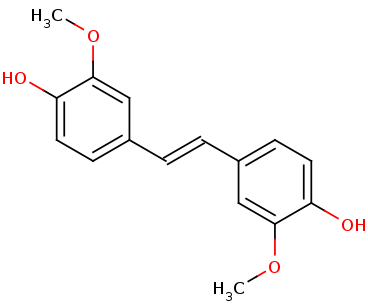
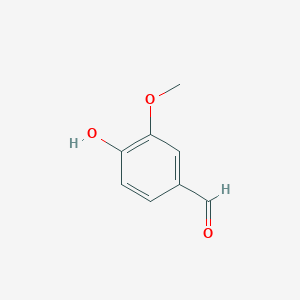
| + | Lignostilbene-α,ß-dioxygenase A (LsdA) from the bacterium ''Sphingomonas paucimobilis'' TMY1009 is a nonheme iron oxygenase that catalyzes the cleavage via oxygenolytic fission of lignostilbene, a compound arising in lignin transformation, to two vanillin molecules (see images below). Lignin is a common component of biomass from industry that scientists are interested in finding ways to break down to simpler and useful materials such as biofuels and commodity chemicals. Though natural occurring lignostilbenoids are rare, they are a very comm on byproduct of industry due to condensation reactions. Other lignin-based stilbenes are thought to be produces from bacterial catabolic processing of diaryl propane and phenylcoumarane <ref>PMID 31292192</ref>. | ||
| - | == Broader Implications == | ||
| - | == Structural highlights and structure-function relationships == | + | ==== '''Lignostilbene''' ==== |
| + | [[Image:lignostilbene.png]] | ||
| + | ==== '''Vanillin''' ==== | ||
| + | [[Image:Vanillin.png]] | ||
| + | == '''Broader Implications''' == | ||
| + | Though lignin is the second most abundant fraction byproduct of lignocellulose, it is currently mostly just combusted for steam and electricity generation. It is rarely valorized into other products like a dispersant for cement and gypsum. One major difficulty in converting and valorizing lignin-based byproducts it their highly variable composition. Both the substrate in which they are located, and the means of extraction affect their end composition and properties. There is much research into finding strong methods of breakdown of lignostilbenoid molecules. | ||
| + | == '''Structural highlights and structure-function relationships''' == | ||
| + | LsdA appears, when crystallized, as two LsdA protomers in one asymmetric unit as a dimer. The <scene name='82/823092/Secondary_and_tertiary_struct/1'>secondary and tertiary structure</scene> of the protomer consists of α-helices (purple) and ß-sheets (blue). The ß-sheets are arranged in a <scene name='82/823092/Seven-bladed_beta_propeller/1'>seven-bladed ß-propeller</scene>, typical of the carotenoid cleavage oxygenases <ref>PMID 31292192</ref>. | ||
| - | = | + | When you look at the <scene name='82/823092/Spacefill_lsda/1'>spacefill view</scene> of the protein dimer you see that the binding pocket accessibility is very restrictive. |
| - | This is a | + | This is a <scene name='82/823092/Hydrophobic_spacefill/1'>Hydrophobicity-focused</scene> view of the protein. Overall, there doesn't seem to be any dominant hydrophobic or hydrophillic regions of the protein. |
| + | |||
| + | The <scene name='82/823092/Catalytic_triad/2'>catalytic triad</scene> of the binding site consists of Phe59, Tyr101, and Lys134 that interact with the 4-hydroxyphenyl portion of the substrate. The triad importance was tested with specific mutations. A F59H mutation led to 3% efficiency comparable to wildtype LsdA. A Y101F mutation led to 20% efficiency comparable to wildtype LsdA. And a K134M mutation showed no discernible lignostilbene cleavage activity <ref>PMID 31292192</ref>. | ||
| + | |||
| + | Important <scene name='82/823092/Active_site_interactions/1'>interactions in the active site</scene> are shown. Green indicates hydrophobic interactions, blue indicates hydrogen bonding interactions, and the orange nucleotides are specific histidines that support and interact with the metal ion (Fe) in the pocket. These two histidines also contribute to hydrogen bonds in the area. | ||
| + | |||
| + | == '''Energy Transformation''' == | ||
| + | |||
| + | LSDs are in the functional protein family called carotenoid cleavage oxygenases (CCOs). They are characterized by their cleavage of the double bond found in carotenoids. The binding site has ferrous iron supported by histidines and is kept in close proximity to the scissile double bond. There are two suggested mechanisms for LSD cleavage. McAndrew ''et al.'' suggested the hydroxystilbenoid is activated by enzyme-catalyzed deprotonation of the 4-hydroxy group. This causes electron delocalization toward the ferrous iron (-superoxo electrophile). The second proposed mechanism suggested by Sui ''et al.'' says that the pi bond electron density from the scissile double bond is redistributed to the iron-oxy complex to form an iron peroxo-substrate cation intermediate. In either case, deprotonation of the hydoxyl group is critical and assisted by the catalytic triad.<ref>PMID 31292192</ref> | ||
| + | |||
| + | The enzyme serves to lower the activation energy and assist in the cleavage and transformation of the lignostilbene to two vanillins. | ||
</StructureSection> | </StructureSection> | ||
| - | == References == | + | == '''References''' == |
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from Aug 26 through Dec 12, 2019 for use in the course CHEM 351 Biochemistry taught by Bonnie_Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1556 through Sandbox Reserved 1575. |
To get started:
More help: Help:Editing |
Lignostilbene-α,ß-dioxygenase A structural features and important functional residues
| |||||||||||
References
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428