We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Nitrite reductase
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
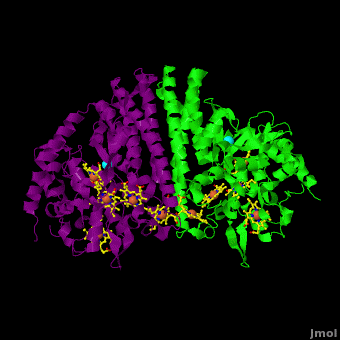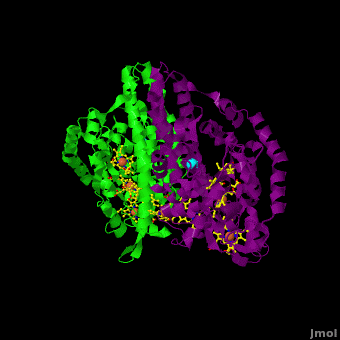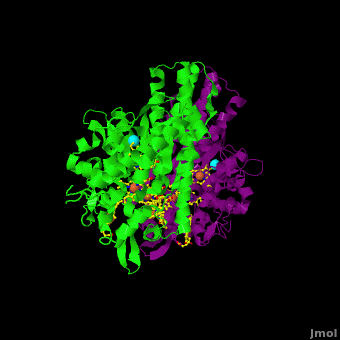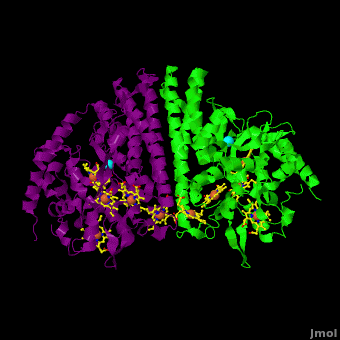
<StructureSection load='Cca.pdb' size='350' side='right' scene='Journal:JBIC:16/Cv/2' caption='Heme-containing nitrite reductase with heme and Ca+2 ions (PDB code [[3ubr]])'> | <StructureSection load='Cca.pdb' size='350' side='right' scene='Journal:JBIC:16/Cv/2' caption='Heme-containing nitrite reductase with heme and Ca+2 ions (PDB code [[3ubr]])'> | ||
| + | |||
| + | __TOC__ | ||
| + | ==Function== | ||
| + | |||
'''Nitrite reductase''' (NIR) catalyzes the reduction of NO<sub>2</sub> to NO. There are 2 classes of NIR: (1) A '''heme-containing cytochrome Cd type NIR'''. This enzyme contains 4 heme groups. Its d-type heme group binds NO<sub>2</sub>. (2) A '''copper-containing NIR''' which produces NO<sub>2</sub>. Under anaerobic conditions bacteria rely on the reduction of nitrogen oxide species to obtain energy. NIR is part of the nitrogen cycle used for this purpose. | '''Nitrite reductase''' (NIR) catalyzes the reduction of NO<sub>2</sub> to NO. There are 2 classes of NIR: (1) A '''heme-containing cytochrome Cd type NIR'''. This enzyme contains 4 heme groups. Its d-type heme group binds NO<sub>2</sub>. (2) A '''copper-containing NIR''' which produces NO<sub>2</sub>. Under anaerobic conditions bacteria rely on the reduction of nitrogen oxide species to obtain energy. NIR is part of the nitrogen cycle used for this purpose. | ||
| Line 5: | Line 9: | ||
For additional details see [[CcNiR]]. | For additional details see [[CcNiR]]. | ||
| - | '''Siroheme-containing NR''' contains siroheme which is a heme-like group used for reduction of sulfur and nitrogen. | + | *'''Siroheme-containing NR''' contains siroheme which is a heme-like group used for reduction of sulfur and nitrogen. |
'''Laue Crystal Structure of ''Shewanella oneidensis'' Cytochrome c Nitrite Reductase from a High-yield Expression System''' <ref name="Youngblut">doi 10.1007/s00775-012-0885-0</ref> | '''Laue Crystal Structure of ''Shewanella oneidensis'' Cytochrome c Nitrite Reductase from a High-yield Expression System''' <ref name="Youngblut">doi 10.1007/s00775-012-0885-0</ref> | ||
| Line 37: | Line 41: | ||
The Ca<sup>2+</sup> ion within <scene name='Journal:JBIC:16/Cv/14'> the conserved site</scene> is coordinated in bidentate fashion by <scene name='Journal:JBIC:16/Cv/15'>Glu205</scene>, and in monodentate fashion by the <scene name='Journal:JBIC:16/Cv/16'>Tyr206 and Lys254</scene> backbone carbonyls, and the <scene name='Journal:JBIC:16/Cv/17'>Gln256</scene> side-chain carbonyl. In the ''S. oneidensis'' structure only <scene name='Journal:JBIC:16/Cv/18'>one water molecule</scene> is assigned to the Ca<sup>2+</sup> ion in subunit B. In subunit A the difference electron density that represents this water molecule is very close to the noise level, and it is difficult to identify even one water molecule there. The <scene name='Journal:JBIC:16/Cv/14'>carbonyl side chain of Asp242 and the hydroxyl of Tyr235</scene> come near to the open calcium coordination sites, but are not within bonding distance. Instead they interact with the water molecule that is weakly coordinated to the Ca<sup>2+</sup> ion. The ccNiR calcium ions appear to play a vital role in organizing the <scene name='Journal:JBIC:16/Cv/13'>active site</scene> (as was mentioned above <font color='magenta'><b>hemes-1</b></font> are the active sites). | The Ca<sup>2+</sup> ion within <scene name='Journal:JBIC:16/Cv/14'> the conserved site</scene> is coordinated in bidentate fashion by <scene name='Journal:JBIC:16/Cv/15'>Glu205</scene>, and in monodentate fashion by the <scene name='Journal:JBIC:16/Cv/16'>Tyr206 and Lys254</scene> backbone carbonyls, and the <scene name='Journal:JBIC:16/Cv/17'>Gln256</scene> side-chain carbonyl. In the ''S. oneidensis'' structure only <scene name='Journal:JBIC:16/Cv/18'>one water molecule</scene> is assigned to the Ca<sup>2+</sup> ion in subunit B. In subunit A the difference electron density that represents this water molecule is very close to the noise level, and it is difficult to identify even one water molecule there. The <scene name='Journal:JBIC:16/Cv/14'>carbonyl side chain of Asp242 and the hydroxyl of Tyr235</scene> come near to the open calcium coordination sites, but are not within bonding distance. Instead they interact with the water molecule that is weakly coordinated to the Ca<sup>2+</sup> ion. The ccNiR calcium ions appear to play a vital role in organizing the <scene name='Journal:JBIC:16/Cv/13'>active site</scene> (as was mentioned above <font color='magenta'><b>hemes-1</b></font> are the active sites). | ||
| - | |||
| - | </StructureSection> | ||
==3D structures of nitrite reductase== | ==3D structures of nitrite reductase== | ||
| + | [[Nitrite reductase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[2akj]] - NIR + S4Fe4 + siroheme – spinach<br /> | ||
| - | **[[3b0g]], [[3b0h]] - ToNIR + S4Fe4 + siroheme – tobacco<br /> | ||
| - | **[[3b0j]], [[3b0l]], [[3b0m]], [[3b0n]], [[3vlx]], [[3vly]], [[3vlz]] - ToNIR (mutant) + S4Fe4 + siroheme <br /> | ||
| - | **[[3vkp]], [[3vkq]], [[3vkr]] – SToNIR + S4Fe4 + siroheme + NO2<br /> | ||
| - | **[[3vm0]] – SToNIR (mutant) + S4Fe4 + siroheme + NO2<br /> | ||
| - | **[[3vm1]] – SToNIR (mutant) + S4Fe4 + siroheme + HCO3<br /> | ||
| - | **[[3vks]] – SToNIR + S4Fe4 + siroheme + NO<br /> | ||
| - | **[[3vkt]] – SToNIR + S4Fe4 + siroheme + NH2OH<br /> | ||
| - | }} | ||
'''References''' | '''References''' | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Youngblut M, Judd ET, Srajer V, Sayyed B, Goelzer T, Elliott SJ, Schmidt M, Pacheco AA. Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. J Biol Inorg Chem. 2012 Mar 2. PMID:22382353 doi:10.1007/s00775-012-0885-0