Phosphofructokinase (PFK)
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 5: | Line 5: | ||
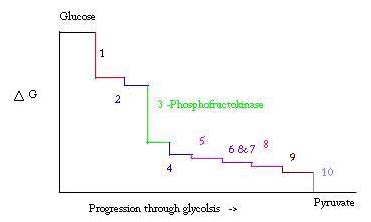
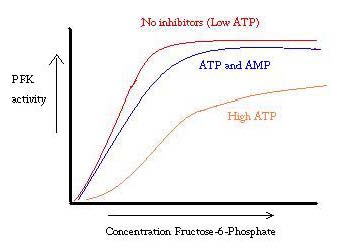
'''Phosphofructokinase-1''' (PFK-1) is a glycolytic enzyme that catalyzes the transfer of a phosphoryl group from <scene name='Phosphofructokinase_(PFK)/Cv/21'>ATP</scene> to <scene name='Phosphofructokinase_(PFK)/Cv/22'>fructose-6-phosphate (F6P)</scene> to yield <scene name='Phosphofructokinase_(PFK)/Cv/16'>ADP</scene> and <scene name='Phosphofructokinase_(PFK)/Cv/23'>fructose-1,6-bisphosphate (FBP)</scene>. See [[Glycolysis Enzymes]]. Mg2+ is also important in this reaction (<scene name='Phosphofructokinase_(PFK)/Cv/24'>click here to see animation of reaction</scene>). '''Phosphofructokinase-2''' (PFK-2) acts on the same substrates to yield ADP and <scene name='Phosphofructokinase_(PFK)/Cv1/3'>fructose-2,6-bisphosphate (F2,6P)</scene>. <scene name='Phosphofructokinase_(PFK)/Cv1/4'>Click here to see the difference between FBP and F2,6P</scene>. PFK reaction is strongly exergonic (irreversible) under physiological conditions and hence is one of the glycolytic pathway's rate-determining steps. In most organisms/tissues, PFK is the glycolytic pathway's major flux-regulating enzyme; its activity is controlled by the concentrations of an unusually large number of metabolites including ATP, ADP, AMP, PEP and fructose-2,6-bisphosphate. | '''Phosphofructokinase-1''' (PFK-1) is a glycolytic enzyme that catalyzes the transfer of a phosphoryl group from <scene name='Phosphofructokinase_(PFK)/Cv/21'>ATP</scene> to <scene name='Phosphofructokinase_(PFK)/Cv/22'>fructose-6-phosphate (F6P)</scene> to yield <scene name='Phosphofructokinase_(PFK)/Cv/16'>ADP</scene> and <scene name='Phosphofructokinase_(PFK)/Cv/23'>fructose-1,6-bisphosphate (FBP)</scene>. See [[Glycolysis Enzymes]]. Mg2+ is also important in this reaction (<scene name='Phosphofructokinase_(PFK)/Cv/24'>click here to see animation of reaction</scene>). '''Phosphofructokinase-2''' (PFK-2) acts on the same substrates to yield ADP and <scene name='Phosphofructokinase_(PFK)/Cv1/3'>fructose-2,6-bisphosphate (F2,6P)</scene>. <scene name='Phosphofructokinase_(PFK)/Cv1/4'>Click here to see the difference between FBP and F2,6P</scene>. PFK reaction is strongly exergonic (irreversible) under physiological conditions and hence is one of the glycolytic pathway's rate-determining steps. In most organisms/tissues, PFK is the glycolytic pathway's major flux-regulating enzyme; its activity is controlled by the concentrations of an unusually large number of metabolites including ATP, ADP, AMP, PEP and fructose-2,6-bisphosphate. | ||
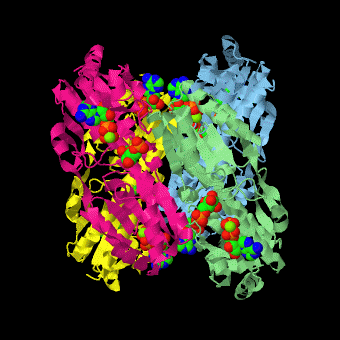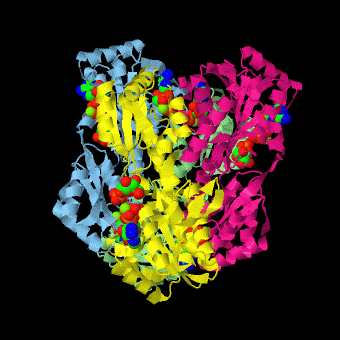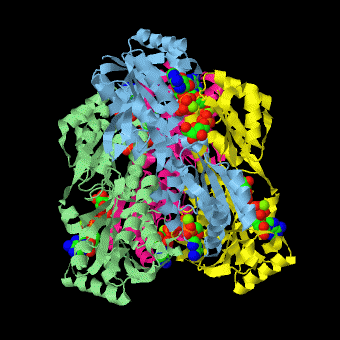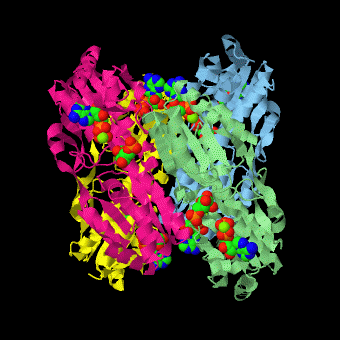
| - | <scene name='Phosphofructokinase_(PFK)/4pfk_biol/3'>PFK from B. stearothermophilus</scene> is a tetramer of identical 320-residue subunits. It is an allosteric enzyme that is described using the symmetry model of allosterism whereby there is a concerted transition from its high-activity R state to its low-activity T state. The X-ray structures of both R and T states of the enzyme have been reported.<ref>PMID:2136935</ref> The binding of one molecule of its substrate F6P, which binds to the R state enzyme with high affinity but to the T state enzyme with low affinity, causes PFK to take up the R state, which in turn, increases the binding affinity of the enzyme for additional F6P (a homotropic effect). Activators, such as ADP and AMP bind to so-called allosteric sites, binding sites distinct from the active site, where they likewise facilitate the formation of the R state and hence activate the enzyme (a heterotropic effect; ADP, being a product of the PFK reaction, also binds at the enzyme's active site). Similarly, inhibitors such as PEP bind to allosteric sites (which in the case of PFK overlaps the activating allosteric site) where they promote the formation of the T state, thereby inhibiting the enzyme. | + | <scene name='Phosphofructokinase_(PFK)/4pfk_biol/3'>PFK from B. stearothermophilus</scene> is a tetramer of identical 320-residue subunits. It is an allosteric enzyme that is described using the symmetry model of allosterism whereby there is a concerted transition from its high-activity R state to its low-activity T state. The X-ray structures of both R and T states of the enzyme have been reported.<ref>PMID:2136935</ref> The binding of one molecule of its substrate <scene name='37/376373/F6p/1'>F6P</scene>, which binds to the R state enzyme with high affinity but to the T state enzyme with low affinity, causes PFK to take up the R state, which in turn, increases the binding affinity of the enzyme for additional F6P (a homotropic effect). Activators, such as <scene name='37/376373/Allosteric_adp/1'>ADP</scene> and AMP bind to so-called allosteric sites, binding sites distinct from the active site, where they likewise facilitate the formation of the R state and hence activate the enzyme (a heterotropic effect; ADP, being a product of the PFK reaction, also binds at the enzyme's active site). Similarly, inhibitors such as PEP bind to allosteric sites (which in the case of PFK overlaps the activating allosteric site) where they promote the formation of the T state, thereby inhibiting the enzyme. |
Two of the active sites of the enzyme are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ad_overview/1'>subunits A (light blue) and D (yellow)</scene> with the active site interfaces in magenta with the substrates in cyan. Two more active sites are at the interface of subunits B (green) and C (pink). A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ad_closeup/1'>A/D interface active site</scene> of subunit D (Yellow) shows that amino acids from both subunits A (light blue) and D (Yellow) contribute to the binding of F6P. Two of the allosteric sites are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ab_overview/1'>subunits A and B</scene> and two at the interface of subunits C and D. Again the interfaces are magenta with the allosteric ligand in cyan. A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ab_closeup/1'>A/B allosteric site</scene> of subunit A shows contributions from both subunits to the binding of ADP. The conformational changes in going between the R and T states of PFK are illustrated below. | Two of the active sites of the enzyme are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ad_overview/1'>subunits A (light blue) and D (yellow)</scene> with the active site interfaces in magenta with the substrates in cyan. Two more active sites are at the interface of subunits B (green) and C (pink). A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ad_closeup/1'>A/D interface active site</scene> of subunit D (Yellow) shows that amino acids from both subunits A (light blue) and D (Yellow) contribute to the binding of F6P. Two of the allosteric sites are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ab_overview/1'>subunits A and B</scene> and two at the interface of subunits C and D. Again the interfaces are magenta with the allosteric ligand in cyan. A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ab_closeup/1'>A/B allosteric site</scene> of subunit A shows contributions from both subunits to the binding of ADP. The conformational changes in going between the R and T states of PFK are illustrated below. | ||
| Line 37: | Line 37: | ||
This kinemage shows the two subunits of the tetramer whose interface contains two active sites. | This kinemage shows the two subunits of the tetramer whose interface contains two active sites. | ||
<kinemage align="right" width="450" height= "450" file="PFK1.kin" /> | <kinemage align="right" width="450" height= "450" file="PFK1.kin" /> | ||
| - | The first view, 1: PFK dimer, shows the two subunits in their R state conformation as represented by their Ca backbones with Subunit 1 in pink tint and Subunit 2 in | + | The first view, 1: <scene name='37/376373/R_161_162/1'>PFK dimer</scene>, shows the two subunits in their R state conformation as represented by their Ca backbones with Subunit 1 in pink tint and Subunit 2 in grey. Two side chains in each subunit are shown, those of Glu 161 (red) and Arg 162 (cyan), which are part of the F6P binding site in the T and R states, respectively(see below). An F6P (hotpink) and an ADP (green; "ADP-active") are bound in the active site of each subunit. An additional ADP (yellow; "ADP-allo") is bound in a separate so-called allosteric site of each subunit. The ADPs each have an associated Mg2<sup>+</sup>, which is represented here by a ball of the same color as the ADP to which it binds. |
Click the "ANIMATE" button to switch the dimer between its R and T states. In its T state, Subunit 1 is bluetint and Subunit 2 is skyblue. The side chains of Glu 161 and Arg 162 in both subunits are red and cyan as before (only the Ca and Cb atoms of the Arg 162 side chain in Subunit 1 are observed in the X-ray structure of the T state; those of Subunit 2 are all observed). The T state enzyme binds the inhibitor 2-phosphoglycolate (gold; "PGC"), a nonphysiological analog of the glycolytic intermediate phosphoenolpyruvate (PEP). Note that the binding site of PGC in the T state overlaps the allosteric binding site of ADP in the R state ("ADP-allo") and hence their binding is mutually exclusive. The T state active sites, which do not contain F6P, are marked by "ghost" F6Ps (gray;"F6P site"), which have the same positions as do the F6Ps in the R state enzyme. | Click the "ANIMATE" button to switch the dimer between its R and T states. In its T state, Subunit 1 is bluetint and Subunit 2 is skyblue. The side chains of Glu 161 and Arg 162 in both subunits are red and cyan as before (only the Ca and Cb atoms of the Arg 162 side chain in Subunit 1 are observed in the X-ray structure of the T state; those of Subunit 2 are all observed). The T state enzyme binds the inhibitor 2-phosphoglycolate (gold; "PGC"), a nonphysiological analog of the glycolytic intermediate phosphoenolpyruvate (PEP). Note that the binding site of PGC in the T state overlaps the allosteric binding site of ADP in the R state ("ADP-allo") and hence their binding is mutually exclusive. The T state active sites, which do not contain F6P, are marked by "ghost" F6Ps (gray;"F6P site"), which have the same positions as do the F6Ps in the R state enzyme. | ||
| Line 53: | Line 53: | ||
KINEMAGE 2 comes up in view 1: The Allosteric Site, in the R state showing the phosphate group of F6P (hotpink) bound in the enzyme's active site in a hydrogen bonded salt bridge (dashed white lines) with the side chain of Arg 162 (cyan). An ADP (yellow; "ADP-allo") occupies the adjacent allosteric site. Click once on "ANIMATE" to switch to the T state. This replaces the ADP in the R state allosteric site with the inhibitor and PEP analog PGC (gold). F6P no longer occupies the active site but its position in the R state is indicated by the "ghost" F6P (gray; viewed by clicking on "F6P site"). | KINEMAGE 2 comes up in view 1: The Allosteric Site, in the R state showing the phosphate group of F6P (hotpink) bound in the enzyme's active site in a hydrogen bonded salt bridge (dashed white lines) with the side chain of Arg 162 (cyan). An ADP (yellow; "ADP-allo") occupies the adjacent allosteric site. Click once on "ANIMATE" to switch to the T state. This replaces the ADP in the R state allosteric site with the inhibitor and PEP analog PGC (gold). F6P no longer occupies the active site but its position in the R state is indicated by the "ghost" F6P (gray; viewed by clicking on "F6P site"). | ||
| - | What happens to the central polypeptide helical segment (residues 149-164) in the R to T transition? What does this do to the relative positions of the negatively charged Glu 161 and the positively charged Arg 162? | + | What happens to the central polypeptide helical segment (residues 149-164) in the R to T transition? <scene name='37/376373/161_162_r/1'>R helix, 161 and 162</scene> <scene name='37/376373/161_162_t/2'>T helix, 161 and 162</scene> What does this do to the relative positions of the negatively charged Glu 161 and the positively charged Arg 162? What influence would the absence of the positive charge of Arg 162 have on the binding of F6P? Does this explain, at least in part, why T state PFK has low affinity for F6P? Go to View 2: Closeup, for a closeup of the F6P-sidechain interactions. Center the molecules by choosing "pickcenter" from the "tools" menu and clicking on athe atom you'd like to be in the center. Slide the "zoom" slider to enlarge the view. |
==Site-Directed Mutagenesis== | ==Site-Directed Mutagenesis== | ||
At one time, the negative charge of Glu 161 was thought to have a negative effect on F6P binding in the T state. This idea has not been supported by site-directed mutagenesis experiments<ref>PMID:10759544</ref>. Several mutant PFKs have been made, including R162A, E161A and R162A/E161A. The R162A mutation caused a 30-fold decrease in F6P binding. The E161A mutation, however, had little effect on the ability of PEP to inhibit F6P binding. | At one time, the negative charge of Glu 161 was thought to have a negative effect on F6P binding in the T state. This idea has not been supported by site-directed mutagenesis experiments<ref>PMID:10759544</ref>. Several mutant PFKs have been made, including R162A, E161A and R162A/E161A. The R162A mutation caused a 30-fold decrease in F6P binding. The E161A mutation, however, had little effect on the ability of PEP to inhibit F6P binding. | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Schirmer T, Evans PR. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990 Jan 11;343(6254):140-5. PMID:2136935 doi:http://dx.doi.org/10.1038/343140a0
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: structure and control. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):53-62. PMID:6115424
- ↑ http://www.nature.com/nature/journal/v327/n6121/abs/327437a0.html
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ PubMed:2136935
- ↑ Campos G, Guixe V, Babul J. Kinetic mechanism of phosphofructokinase-2 from Escherichia coli. A mutant enzyme with a different mechanism. J Biol Chem. 1984 May 25;259(10):6147-52. PMID:6233271
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ PubMed:2136935
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Campos G, Guixe V, Babul J. Kinetic mechanism of phosphofructokinase-2 from Escherichia coli. A mutant enzyme with a different mechanism. J Biol Chem. 1984 May 25;259(10):6147-52. PMID:6233271
- ↑ Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):3844-9. PMID:10759544 doi:10.1073/pnas.050588097
- ↑ Vora S, Corash L, Engel WK, Durham S, Seaman C, Piomelli S. The molecular mechanism of the inherited phosphofructokinase deficiency associated with hemolysis and myopathy. Blood. 1980 Apr;55(4):629-35. PMID:6444532
External Links
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Judy Voet, Ann Taylor, Jaime Prilusky, David Canner, Eran Hodis, Joel L. Sussman, Zach Westrick