|
|
| (One intermediate revision not shown.) |
| Line 3: |
Line 3: |
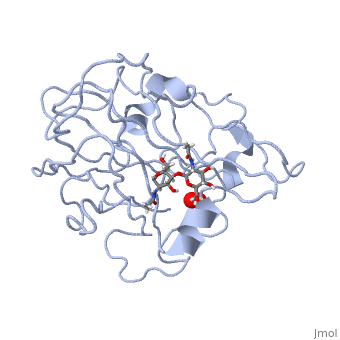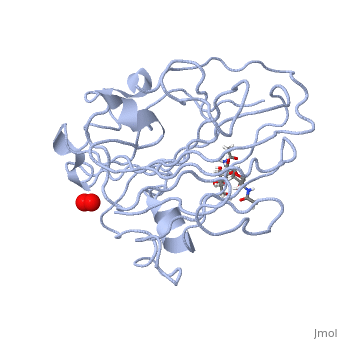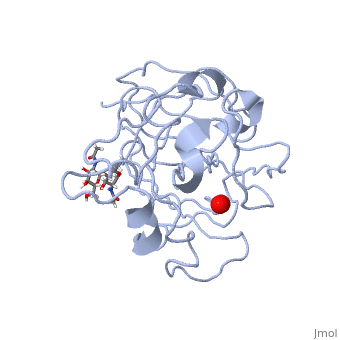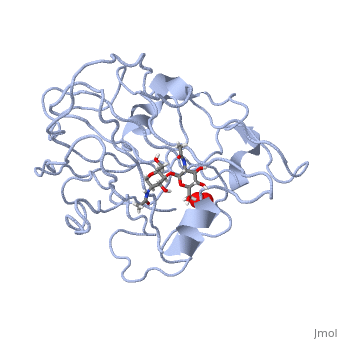
| | <StructureSection load='5tkh' size='340' side='right'caption='[[5tkh]], [[Resolution|resolution]] 1.20Å' scene=''> | | <StructureSection load='5tkh' size='340' side='right'caption='[[5tkh]], [[Resolution|resolution]] 1.20Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[5tkh]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Chrysonilia_crassa Chrysonilia crassa]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5TKH OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5TKH FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[5tkh]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Neurospora_crassa Neurospora crassa]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5TKH OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=5TKH FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=CU:COPPER+(II)+ION'>CU</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=OXY:OXYGEN+MOLECULE'>OXY</scene>, <scene name='pdbligand=PER:PEROXIDE+ION'>PER</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.2Å</td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[5tki|5tki]], [[5tkf|5tkf]], [[5tkg|5tkg]]</td></tr>
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CU:COPPER+(II)+ION'>CU</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=OXY:OXYGEN+MOLECULE'>OXY</scene>, <scene name='pdbligand=PER:PEROXIDE+ION'>PER</scene></td></tr> |
| - | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">G15G9.090, GE21DRAFT_7469 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=5141 Chrysonilia crassa])</td></tr>
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=5tkh FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5tkh OCA], [https://pdbe.org/5tkh PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=5tkh RCSB], [https://www.ebi.ac.uk/pdbsum/5tkh PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=5tkh ProSAT]</span></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5tkh FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5tkh OCA], [http://pdbe.org/5tkh PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=5tkh RCSB], [http://www.ebi.ac.uk/pdbsum/5tkh PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=5tkh ProSAT]</span></td></tr> | + | |
| | </table> | | </table> |
| | + | == Function == |
| | + | [https://www.uniprot.org/uniprot/LPMO_NEUCR LPMO_NEUCR] Catalyzes the oxidative cleavage of glycosidic bonds in cellulosic substrates via a copper-dependent mechanism (PubMed:22004347, PubMed:22188218, PubMed:24350607, PubMed:31431506). In the presence of an exogenous reductant ascorbic acid, degrades phosphoric acid swollen cellulose (PASC) to cello-oligosaccharides and 4-ketoaldoses, the end products oxidized at the non-reducing end (PubMed:22004347, PubMed:22188218, PubMed:24350607). Somewhat active toward tamarind xyloglucan and konjac glucomannan, with improved activity with glucomannan in the presence of PASC (PubMed:31431506). H(2)O(2) is able to substitute for O(2) in reactions with PASC, xyloglucan and glucomannan (PubMed:31431506). Very weak activity on cellopentaose (PubMed:31431506). No activity with birchwood xylan or ivory nut mannan (PubMed:31431506). Disrupts plant cell wall polysaccharide substrates, such as recalcitrant crystalline cellulose (Probable).<ref>PMID:22004347</ref> <ref>PMID:22188218</ref> <ref>PMID:24350607</ref> <ref>PMID:31431506</ref> |
| | <div style="background-color:#fffaf0;"> | | <div style="background-color:#fffaf0;"> |
| | == Publication Abstract from PubMed == | | == Publication Abstract from PubMed == |
| Line 26: |
Line 27: |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Chrysonilia crassa]] | |
| | [[Category: Large Structures]] | | [[Category: Large Structures]] |
| - | [[Category: Dell, W B.O]] | + | [[Category: Neurospora crassa]] |
| - | [[Category: Meilleur, F]] | + | [[Category: Meilleur F]] |
| - | [[Category: Oxidoreductase]] | + | [[Category: O'Dell WB]] |
| - | [[Category: Polysaccharide monooxygenase]]
| + | |
| Structural highlights
Function
LPMO_NEUCR Catalyzes the oxidative cleavage of glycosidic bonds in cellulosic substrates via a copper-dependent mechanism (PubMed:22004347, PubMed:22188218, PubMed:24350607, PubMed:31431506). In the presence of an exogenous reductant ascorbic acid, degrades phosphoric acid swollen cellulose (PASC) to cello-oligosaccharides and 4-ketoaldoses, the end products oxidized at the non-reducing end (PubMed:22004347, PubMed:22188218, PubMed:24350607). Somewhat active toward tamarind xyloglucan and konjac glucomannan, with improved activity with glucomannan in the presence of PASC (PubMed:31431506). H(2)O(2) is able to substitute for O(2) in reactions with PASC, xyloglucan and glucomannan (PubMed:31431506). Very weak activity on cellopentaose (PubMed:31431506). No activity with birchwood xylan or ivory nut mannan (PubMed:31431506). Disrupts plant cell wall polysaccharide substrates, such as recalcitrant crystalline cellulose (Probable).[1] [2] [3] [4]
Publication Abstract from PubMed
Lytic polysaccharide monooxygenases have attracted vast attention owing to their abilities to disrupt glycosidic bonds via oxidation instead of hydrolysis and to enhance enzymatic digestion of recalcitrant substrates including chitin and cellulose. We have determined high-resolution X-ray crystal structures of an enzyme from Neurospora crassa in the resting state and of a copper(II) dioxo intermediate complex formed in the absence of substrate. X-ray crystal structures also revealed "pre-bound" molecular oxygen adjacent to the active site. An examination of protonation states enabled by neutron crystallography and density functional theory calculations identified a role for a conserved histidine in promoting oxygen activation. These results provide a new structural description of oxygen activation by substrate free lytic polysaccharide monooxygenases and provide insights that can be extended to reactivity in the enzyme-substrate complex.
Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.,O'Dell WB, Agarwal PK, Meilleur F Angew Chem Int Ed Engl. 2017 Jan 16;56(3):767-770. doi: 10.1002/anie.201610502., Epub 2016 Dec 22. PMID:28004877[5]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011 Dec 16;6(12):1399-406. PMID:22004347 doi:10.1021/cb200351y
- ↑ Beeson WT, Phillips CM, Cate JH, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012 Jan 18;134(2):890-2. PMID:22188218 doi:10.1021/ja210657t
- ↑ Vu VV, Beeson WT, Phillips CM, Cate JH, Marletta MA. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc. 2014 Jan 15;136(2):562-5. PMID:24350607 doi:10.1021/ja409384b
- ↑ Petrovic DM, Varnai A, Dimarogona M, Mathiesen G, Sandgren M, Westereng B, Eijsink VGH. Comparison of three seemingly similar lytic polysaccharide monooxygenases from Neurospora crassa suggests different roles in plant biomass degradation. J Biol Chem. 2019 Oct 11;294(41):15068-15081. doi: 10.1074/jbc.RA119.008196. Epub, 2019 Aug 20. PMID:31431506 doi:http://dx.doi.org/10.1074/jbc.RA119.008196
- ↑ O'Dell WB, Agarwal PK, Meilleur F. Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase. Angew Chem Int Ed Engl. 2017 Jan 16;56(3):767-770. doi: 10.1002/anie.201610502., Epub 2016 Dec 22. PMID:28004877 doi:http://dx.doi.org/10.1002/anie.201610502
|