We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Retroviral Integrase
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
==Function== | ==Function== | ||
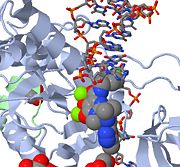
| - | [[Retroviral Integrase]] is an essential retroviral enzyme that binds to viral DNA and inserts it into a host cell chromosome. The reverse transcribed cDNA of human immunodeficiency virus type 1 (HIV-1) is inserted in the host cell genome in order increase pathogen fitness and virulence. Integrase is produced by a class of retrovirus (like HIV) and is used by the virus to incorporate its genetic material into the host cell DNA. The host cellular machinery then produces mRNA and then protein from the incorporated genetic material, thus replicating the virus. Although several integrase inhibiting drugs have been investigated, the mechanism responsible for strand-transfer inhibition action remains to be elucidated <ref>PMID:19915684</ref>. However, Hare el al (2010)<ref name=Hare_2010>PMID:20118915</ref> determined the structural constituents of retroviral integration. Further elucidation of the complete structure of the retroviral integrase, and its application to regulate functional and enzymatic activities could potentially enable researchers to delay the progression of retroviral diseases. Moreover, study of HIV-1 integration could lead to a promising new target, and contribute to the generation pharmacophore models for antiviral therapy<ref>deJesus, Edwin [http://www.thebody.com/content/art1352.html#ii HIV Antiretroviral Agents in Development]. The Body: The Complete HIV/AIDS Resource. March 30, 2006</ref>. <br/> | + | [[Retroviral Integrase]] is an essential retroviral enzyme that binds to viral DNA and inserts it into a host cell chromosome. The complex of integrase and DNA is called '''intasome.''' The reverse transcribed cDNA of human immunodeficiency virus type 1 (HIV-1) is inserted in the host cell genome in order increase pathogen fitness and virulence. Integrase is produced by a class of retrovirus (like HIV) and is used by the virus to incorporate its genetic material into the host cell DNA. The host cellular machinery then produces mRNA and then protein from the incorporated genetic material, thus replicating the virus. Although several integrase inhibiting drugs have been investigated, the mechanism responsible for strand-transfer inhibition action remains to be elucidated <ref>PMID:19915684</ref>. However, Hare el al (2010)<ref name=Hare_2010>PMID:20118915</ref> determined the structural constituents of retroviral integration. Further elucidation of the complete structure of the retroviral integrase, and its application to regulate functional and enzymatic activities could potentially enable researchers to delay the progression of retroviral diseases. Moreover, study of HIV-1 integration could lead to a promising new target, and contribute to the generation pharmacophore models for antiviral therapy<ref>deJesus, Edwin [http://www.thebody.com/content/art1352.html#ii HIV Antiretroviral Agents in Development]. The Body: The Complete HIV/AIDS Resource. March 30, 2006</ref>. <br/> |
HIV Integrase inhibitors: [http://www.isentress.com Raltegravir], marketed as Isentress is currently approved as a therapeutic inhibitor of HIV integrase. It was approved on October 12, 2007. | HIV Integrase inhibitors: [http://www.isentress.com Raltegravir], marketed as Isentress is currently approved as a therapeutic inhibitor of HIV integrase. It was approved on October 12, 2007. | ||
[See below for a table of antiretroviral drugs with trade name, company, patents, and notes.] | [See below for a table of antiretroviral drugs with trade name, company, patents, and notes.] | ||
| Line 19: | Line 19: | ||
[[Image:HIV_Stats.jpg|thumb|alt=Alt text|In 2010, there are more than 25 million people have died of AIDS, and it is estimated that approximately 33 million people are living with HIV.]] | [[Image:HIV_Stats.jpg|thumb|alt=Alt text|In 2010, there are more than 25 million people have died of AIDS, and it is estimated that approximately 33 million people are living with HIV.]] | ||
| - | To date, more than 25 million people have died of AIDS and it is estimated that approximately 33 million people are living with HIV today<ref> [https://aidsinfo.nih.gov AIDS-Info]</ref>. For retroviral integrase inhibitors see [[Raltegravir]] and [[Retroviral Integrase Inhibitor Pharmacokinetics]]. | + | To date, more than 25 million people have died of AIDS and it is estimated that approximately 33 million people are living with HIV today<ref> [https://aidsinfo.nih.gov AIDS-Info]</ref>. For retroviral integrase inhibitors see [[Raltegravir]], [[Genvoya]] and [[Retroviral Integrase Inhibitor Pharmacokinetics]]. |
==Impact of Structure== | ==Impact of Structure== | ||
| Line 87: | Line 87: | ||
</StructureSection> | </StructureSection> | ||
| - | == 3D Structures of Retroviral Integrase == | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Human spumaretrovirus integrase | ||
| - | |||
| - | **[[3os0]], [[3os1]], [[3os2]], [[3oy9]], [[3l2q]], [[3l2r]], [[4e7j]], [[4e7k]], [[4e7l]], [[5uoq]], [[5uop]], [[5no1]], [[5mmb]], [[5mma]], [[5fro]], [[5frm]], [[4ztj]], [[4ztf]] – IN + DNA – human spumaretrovirus<br /> | ||
| - | **[[3oyb]], [[3oyc]], [[3oyd]], [[3oye]], [[3oyf]], [[3oyg]], [[3oyh]], [[3oyi]], [[3oyj]], [[3oyk]], [[3oyl]], [[3oym]], [[3oyn]], [[3oya]], [[3l2u]], [[3l2v]], [[3l2w]] – IN + DNA + inhibitor<br /> | ||
| - | **[[2x6n]], [[2x6s]], [[2x74]], [[2x78]], [[3dlr]] - IN catalytic core domain | ||
| - | |||
| - | *Bovine immunodeficiency virus integrase | ||
| - | |||
| - | **[[3kkr]], [[3kks]] – IN catalytic core domain – BIV | ||
| - | |||
| - | *Human immunodeficiency virus 1 integrase | ||
| - | |||
| - | **[[5oi5]], [[5oi3]], [[5oi2]], [[1itg]], [[6jcg]], [[6jcf]] – IN catalytic core domain <br /> | ||
| - | **[[3lpt]], [[3lpu]], [[3l3u]], [[1exq]], [[1b92]], [[1b9d]], [[1b9f]], [[1bi4]], [[1bhl]], [[1bl3]], [[1bis]], [[1biu]], [[1biz]], [[2itg]], [[3zcm]], [[5jl4]] - IN catalytic core domain (mutant) – HIV-1<br /> | ||
| - | **[[3l3v ]]- IN catalytic core domain (mutant) + sucrose<br /> | ||
| - | **[[1hyv]], [[1hyz]], [[1qs4]] - IN N-terminal + inhibitor<br /> | ||
| - | **[[1k6y]] - IN N-terminal + catalytic core domain<br /> | ||
| - | **[[1wje]], [[1wjf]] - IN N-terminal zinc-binding domain (mutant) – NMR<br /> | ||
| - | **[[1wja]], [[1wjb]], [[1wjc]], [[1wjd]] - IN N-terminal zinc-binding domain<br /> | ||
| - | **[[1ex4]] - IN C-terminal DBD + catalytic core domain (mutant)<br /> | ||
| - | **[[1ihv]], [[1ihw]] – IN DBD - NMR<br /> | ||
| - | **[[1qmc]] – IN DBD - NMR<br /> | ||
| - | **[[2b4j]], [[3av9]], [[3ava]], [[3avb]], [[3avc]], [[3avf]], [[3avg]], [[3avh]], [[3avi]], [[3avj]], [[3avk]], [[3avl]], [[3avm]], [[3avn]] - IN (mutant) + lens epithelium-derived growth factor <br /> | ||
| - | **[[6ex9]], [[5krt]], [[5krs]], [[5kgx]], [[5kgw]], [[4o0j]] – IN catalytic core domain + inhibitor <br /> | ||
| - | **[[3nf6]], [[3nf7]], [[3nf8]], [[3nf9]], [[3nfa]], [[4e1m]], [[4e1n]], [[4cj4]], [[4jlh]], [[4ojr]], [[5hrs]], [[5hrr]], [[5hrp]], [[5hrn]], [[5hot]], [[4tsx]], [[4ovl]], [[4ojr]], [[4o5b]], [[4o55]], [[4nyf]], [[6nuj]], [[6ncj]], [[6eb2]], [[6eb1]] - IN core domain (mutant) + inhibitor<br /> | ||
| - | **[[5tc2]] – IN C terminal <br /> | ||
| - | **[[5eu7]] – IN catalytic core domain + antibody <br /> | ||
| - | **[[4y1d]], [[4y1c]] – IN catalytic core domain + peptide <br /> | ||
| - | |||
| - | *Human immunodeficiency virus 2 integrase | ||
| - | |||
| - | **[[3f9k]] - IN N-terminal + catalytic core domain + LEDGF IBD – HIV-2<br /> | ||
| - | **[[1e0e]] - IN N-terminal – NMR | ||
| - | |||
| - | *Moloney murine leukemia virus integrase | ||
| - | |||
| - | **[[3nnq]], [[4nzg]] – IN N-terminal – MOMLV | ||
| - | |||
| - | *Maedi Visna virus integrase | ||
| - | |||
| - | **[[3hpg]], [[3hph]] – IN N-terminal + catalytic core domain + LEDGF IBD – MVV<br /> | ||
| - | **[[5t3a]] – IN <br /> | ||
| - | **[[5llj]] – IN C terminal<br /> | ||
| - | **[[5m0r]], [[5m0q]] – IN + DNA – Cryo EM<br /> | ||
| - | |||
| - | *Simian immunodeficiency virus integrase | ||
| - | |||
| - | **[[1c6v]] – IN catalytic domain + DNA binding domain - SIV | ||
| - | |||
| - | *Feline immunodeficiency virus integrase | ||
| - | |||
| - | **[[4pa1]] - IN catalytic core domain - FIV<br /> | ||
| - | **[[4mq3]] – IN catalytic core domain (mutant)<br /> | ||
| - | |||
| - | *Mouse mammary tumor virus | ||
| - | |||
| - | **[[5cz1]] – IN catalytic domain <br /> | ||
| - | |||
| - | *Rous sarcoma virus integrase | ||
| - | |||
| - | **[[1c0m]], [[1c1a]] – IN fragment (mutant) – RSV<br /> | ||
| - | **[[1vsk]], [[1vsl]], [[3o4q]] - IN catalytic core domain (mutant)<br /> | ||
| - | **[[1vsm]], [[1vsh]], [[1vsi]], [[1vsj]], [[1vsd]], [[1vse]], [[1vsf]], [[3o4n]] - IN catalytic core domain<br /> | ||
| - | **[[1a5v]], [[1a5w]], [[1a5x]] - IN catalytic core domain + inhibitor<br /> | ||
| - | **[[5ejk]] – IN + DNA + gag-pro-pol polyprotein <br /> | ||
| - | |||
| - | *Avian sarcoma virus integrase | ||
| - | |||
| - | **[[1cxq]], [[1cxu]], [[1cz9]], [[1czb]], [[1asu]], [[1asv]], [[1asw]] - IN catalytic core domain (mutant) – ASV | ||
| - | |||
| - | *Bacterial integrase | ||
| - | |||
| - | **[[3nkh]] – IN fragment – ''Staphylococcus aureus'' MRSA<br /> | ||
| - | **[[1b69]], [[1tn9]] - EfIN N-terminal DBD + DNA – ''Enterococcus faecalis'' – NMR<br /> | ||
| - | **[[1bb8]], [[2bb8]] - EfIN N-terminal DBD – NMR<br /> | ||
| - | **[[3nrw]] – IN/site-specific recombinase N-terminal – ''Haloarcula marismortui''<br /> | ||
| - | **[[3jtz]] – IN arm-type binding domain – ''Yersinia pestis''<br /> | ||
| - | **[[3ju0]] – HPI IN arm-type binding domain – ''Pectobacterium atrosepticum''<br /> | ||
| - | **[[2kkp]] – IN SAM-like domain – ''Moorella thermoacetica'' – NMR<br /> | ||
| - | **[[2kkv]] – IN fragment – ''Salmonella enterica'' – NMR<br /> | ||
| - | **[[2khq]] - IN fragment – ''Staphylococcus saprophyticus'' – NMR<br /> | ||
| - | **[[2khv]], [[2kj5]] – INT residues 102-199 – ''Nitrosospira multiformis'' – NMR<br /> | ||
| - | **[[2a3v]] – Rec INTI4+DNA – ''Vibrio cholera''<br /> | ||
| - | **[[1z19]], [[1z1b]], [[1z1g]], [[1p7d]] – EpLamInt catalytic core domain+DNA<br /> | ||
| - | **[[6emy]], [[6emz]], [[6en0]], [[6en1]], [[6en2]] – IN (mutant) + DNA – Enterococcus faecalis<br /> | ||
| - | |||
| - | *Phage integrase | ||
| - | |||
| - | **[[3bvp]] – Integrase N-terminal – Lactococcus phage <br /> | ||
| - | **[[1aih]] – HP1 integrase catalytic domain – bacteriophage<br /> | ||
| - | **[[5dor]], [[5c6k]] – IN catalytic domain – Enterobacteria phage P2 <br /> | ||
| - | **[[2wcc]], [[1kjk]] – λIN DBD+ DNA – Enterobacteria phage λ - NMR<br /> | ||
| - | **[[2oxo]] – λIN residues 75-176<br /> | ||
| - | **[[1ae9]] – λIN catalytic core<br /> | ||
| - | **[[5j0n]] – λIN + ATTP + ATTB + integration host factor + excisionase - Cryo EM<br /> | ||
| - | **[[5udo]] – IN coiled-coil domain – Listeria innocua phage <br /> | ||
| - | |||
| - | *Yeast integrase | ||
| - | |||
| - | **[[1p4e]], 1m6x]] – FLP (mutant)+DNA <br /> | ||
| - | **[[1flo]] - FLP +DNA<br /> | ||
| - | }} | ||
| - | |||
==References== | ==References== | ||
Current revision
| |||||||||||
References
- ↑ Pandey KK, Grandgenett DP. HIV-1 Integrase Strand Transfer Inhibitors: Novel Insights into their Mechanism of Action. Retrovirology (Auckl). 2008 Nov 5;2:11-16. PMID:19915684
- ↑ 2.0 2.1 2.2 2.3 Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010 Mar 11;464(7286):232-6. Epub 2010 Jan 31. PMID:20118915 doi:10.1038/nature08784
- ↑ deJesus, Edwin HIV Antiretroviral Agents in Development. The Body: The Complete HIV/AIDS Resource. March 30, 2006
- ↑ Braun, J.F., Cronje, R.J and Henderson, M.G. HIV-1 Integrase Inhibitors Inhibitors (2008). www.prn.org Volume 13, Pages 1–9
- ↑ AIDS-Info
- ↑ Iwamoto M, Wenning LA, Petry AS, Laethem M, De Smet M, Kost JT, Breidinger SA, Mangin EC, Azrolan N, Greenberg HE, Haazen W, Stone JA, Gottesdiener KM, Wagner JA. Minimal effects of ritonavir and efavirenz on the pharmacokinetics of raltegravir. Antimicrob Agents Chemother. 2008 Dec;52(12):4338-43. Epub 2008 Oct 6. PMID:18838589 doi:10.1128/AAC.01543-07
Further Reading
- GEN News Highlights "Scientists Solve 3-D Crystal Structure of Retroviral Integrase Bound to Viral DNA", Genetic Engineering & Biotechnology News February 1, 2010.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Rhysly Martinez, Joel L. Sussman, Alexander Berchansky, David Canner, Jordan Heard, Eugene Babcock, Garrett Asanuma