This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
LdtMt2
From Proteopedia
| (5 intermediate revisions not shown.) | |||
| Line 37: | Line 37: | ||
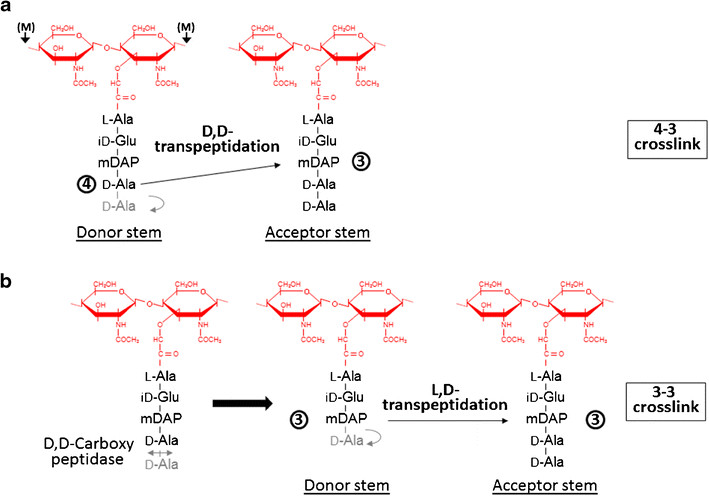
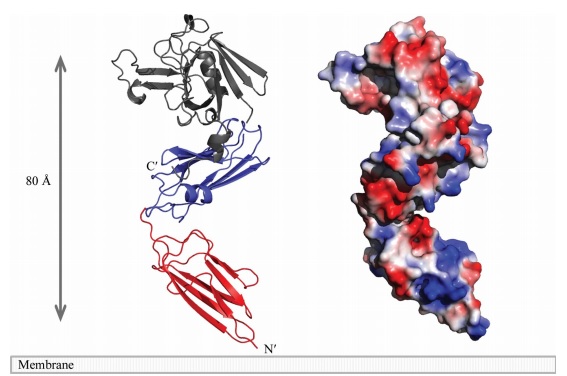
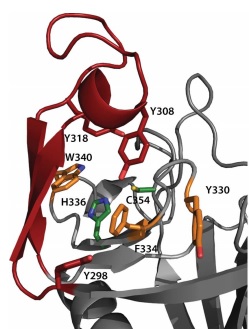
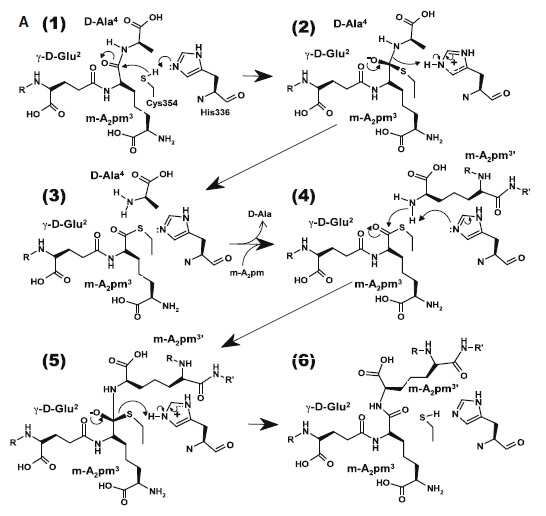
At one end of the tunnel <scene name='81/817533/D-chiral_center_recognition/2'>Tyr318 and the carbonyl group of Gly332</scene> recognize the D chiral center of the meso-diaminopimelic acid side chain, making H bonds to its carboxylate and amide group. At the other end, the L chiral center is surrounded by an <scene name='81/817533/Anion_hole/1'>anion hole</scene> formed by the backbone NH group of residues 352–354 at the C terminus of loop Lc. In this position, the acyl group of the m-DAP L chiral center is within reach of Cys354. '''His336''' is close and ready to accept a H+ from '''Cys354''' (<scene name='81/817533/Conserved_residues/1'>conserved residues</scene>). The cysteine thiolate formed by this H+ abstraction attacks the acyl carbon and forms a tetrahedral intermediate stabilized by the ‘‘anion hole’’ and by the just protonated '''His336'''. After the intermediate thioester formation and protonation by '''His336''', D-Ala4 is released. Then another peptide stem can enter the catalytic site and bind to active site residues with the side-chain amide of the mDAP residue. Nucleophilic attack by this amine group forms the new peptide bond that crosslinks the two stems with release and protonation of the cysteine thiolate by '''His336'''. The last step in the transpeptidation reaction requires that donor and acceptor peptidoglycan stems both be at the catalytic site for the reaction to occur. | At one end of the tunnel <scene name='81/817533/D-chiral_center_recognition/2'>Tyr318 and the carbonyl group of Gly332</scene> recognize the D chiral center of the meso-diaminopimelic acid side chain, making H bonds to its carboxylate and amide group. At the other end, the L chiral center is surrounded by an <scene name='81/817533/Anion_hole/1'>anion hole</scene> formed by the backbone NH group of residues 352–354 at the C terminus of loop Lc. In this position, the acyl group of the m-DAP L chiral center is within reach of Cys354. '''His336''' is close and ready to accept a H+ from '''Cys354''' (<scene name='81/817533/Conserved_residues/1'>conserved residues</scene>). The cysteine thiolate formed by this H+ abstraction attacks the acyl carbon and forms a tetrahedral intermediate stabilized by the ‘‘anion hole’’ and by the just protonated '''His336'''. After the intermediate thioester formation and protonation by '''His336''', D-Ala4 is released. Then another peptide stem can enter the catalytic site and bind to active site residues with the side-chain amide of the mDAP residue. Nucleophilic attack by this amine group forms the new peptide bond that crosslinks the two stems with release and protonation of the cysteine thiolate by '''His336'''. The last step in the transpeptidation reaction requires that donor and acceptor peptidoglycan stems both be at the catalytic site for the reaction to occur. | ||
| + | |||
| + | </StructureSection> | ||
==3D structures of L,D-transpeptidase== | ==3D structures of L,D-transpeptidase== | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
*L,D-transpeptidase | *L,D-transpeptidase | ||
| Line 58: | Line 61: | ||
**[[3vyo]], [[4huc]], [[3tur]], [[3u1p]], [[3u1q]], [[3vae]] – MtLDT 2 B and C domain residues 140-408<br /> | **[[3vyo]], [[4huc]], [[3tur]], [[3u1p]], [[3u1q]], [[3vae]] – MtLDT 2 B and C domain residues 140-408<br /> | ||
**[[3tx4]] – MtLDT 2 B and C domain (mutant)<br /> | **[[3tx4]] – MtLDT 2 B and C domain (mutant)<br /> | ||
| - | **5d7h]], [[5du7]] – MtLDT 2 residues 56-407<br /> | + | **[[5d7h]], [[5du7]] – MtLDT 2 residues 56-407<br /> |
| - | **[6d4k]] – MtLDT 3 A and B domain <br /> | + | **[[6d4k]] – MtLDT 3 A and B domain <br /> |
**[[4z7a]], [[6d5a]] – MtLDT 5<br /> | **[[4z7a]], [[6d5a]] – MtLDT 5<br /> | ||
| Line 66: | Line 69: | ||
**[[5e51]], [[4jmx]] – MtLDT 1 A and B domain + antibiotic <br /> | **[[5e51]], [[4jmx]] – MtLDT 1 A and B domain + antibiotic <br /> | ||
**[[5dvp]], [[5dc2]], [[5dcc]], [[5dzj]], [[5dzp]], [[5e1g]], [[5e1i]], [[5k69]], [[6boi]], [[5duj]] – MtLDT 2 residues 56-408 + antibiotic <br /> | **[[5dvp]], [[5dc2]], [[5dcc]], [[5dzj]], [[5dzp]], [[5e1g]], [[5e1i]], [[5k69]], [[6boi]], [[5duj]] – MtLDT 2 residues 56-408 + antibiotic <br /> | ||
| - | **[[3vyp]], [[4gsq]], [[4gsr]], [[4gsu]], [[ | + | **[[3vyp]], [[4gsq]], [[4gsr]], [[4gsu]], [[6iyv]], [[6iyw]], [[5lbg]] – MtLDT 2 B and C domain + antibiotic<br /> |
**[[5lb1]] – MtLDT 2 B and C domain + thionitrobenzoate<br /> | **[[5lb1]] – MtLDT 2 B and C domain + thionitrobenzoate<br /> | ||
**[[6d51]] – MtLDT 3 A and B domain + antibiotic<br /> | **[[6d51]] – MtLDT 3 A and B domain + antibiotic<br /> | ||
| Line 91: | Line 94: | ||
<references/> | <references/> | ||
| - | |||
| - | </StructureSection> | ||
Current revision
| |||||||||||
3D structures of L,D-transpeptidase
Updated on 16-January-2020
References
Böth, D., Steiner, E. M., Stadler, D., Lindqvist, Y., Schnell, R., & Schneider, G. (2013). Structure of LdtMt2, an L, D-transpeptidase from Mycobacterium tuberculosis. Acta Crystallographica Section D: Biological Crystallography, 69(3), 432-441.[1]
Brammer, L. B., Ghosh, A., Pan, Y., Jakoncic, J., Lloyd, E. P., Townsend, C. A., ... & Bianchet, M. A. (2015). Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis. The Journal of biological chemistry, 290(42), 25670-25685.[2]
Erdemli, S. B., Gupta, R., Bishai, W. R., Lamichhane, G., Amzel, L. M., & Bianchet, M. A. (2012). Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of L, D-transpeptidase 2. Structure, 20(12), 2103-2115. [3]
Li, W. J., Li, D. F., Hu, Y. L., Zhang, X. E., Bi, L. J., & Wang, D. C. (2013). Crystal structure of L, D-transpeptidase Ldt Mt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis. Cell research, 23(5), 728 [4]
Mainardi, J. L., Fourgeaud, M., Hugonnet, J. E., Dubost, L., Brouard, J. P., Ouazzani, J., ... & Arthur, M. (2005). A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. Journal of Biological Chemistry, 280(46), 38146-38152. [5]