We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Carson E Middlebrook/Sandbox 1
From Proteopedia
< User:Carson E Middlebrook(Difference between revisions)
(New page: ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> This is a default text for you...) |
|||
| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | = | + | =Bd Oxidase from ''Geobacillus thermodenitrificans''= |
| - | <StructureSection load=' | + | <StructureSection load='5DOQ' size='350' frame='true' side='right' caption='Bd Oxidase from Geobacillus thermodenitrificans 5DOQ' scene=’'83/838655/Bdoxidase_structure_full/1'> |
| - | + | ||
| - | + | ||
| + | == Introduction == | ||
| + | |||
| + | <scene name='83/838655/Bdoxidase_structure_full/2'>TextToBeDisplayed</scene> | ||
== Function == | == Function == | ||
| - | == | + | == Active Site == |
| - | = | + | The active site for Bd Oxidase in ''Geobacillus thermodenitrificans'' is located in subunit Cyd A. The site consists of three iron hemes: Heme B558, Heme B595, and Heme D that are held together in a rigid triangular <scene name='83/838655/Hemes/1'>arrangement</scene> due to van der wals interactions. The length between each heme's central iron is relatively constant which serves to shuttle protons and electrons from one heme to another efficiently. It is suggested that Heme B558 acts as an electron acceptor to the extracellular side and Heme B559 acts as a proton acceptor on the intracellular side. It is then proposed that both heme B558 and B595 shuttle their respective ions directly to Heme D based on this being the shortest pathway (reference). Heme D is then suggested to be the oxygen binding site due to proximity and orientation to the exterior surface of the protein. |
| + | == Relevance == | ||
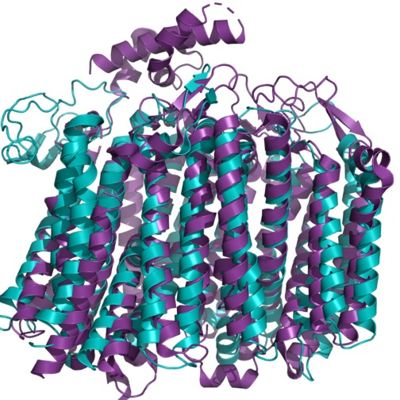
| + | [[Image:Aligmentbdoidase.jpg|400 px|right|thumb|Figure 1. The coolest image of this protein!!!]] | ||
== Structural highlights == | == Structural highlights == | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | + | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. Scene created by Carson: <scene name='83/838655/Residues_30_65/1'>Residues 30-65</scene> <ref name=”Ransey”>PMID:27126043 </ref>. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | == Student Contributers == | ||
| + | *Carson Middlebrook | ||
| + | *Emma Harris | ||
Current revision
Bd Oxidase from Geobacillus thermodenitrificans
| |||||||||||
References
- ↑ Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
Student Contributers
- Carson Middlebrook
- Emma Harris