|
|
| (558 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | <StructureSection load='6vsb' size='250' side='right' scene='83/839266/Covid-19_spike/6' caption='COVID-19 surface spike (PDB entry [[6vsb]])'> | + | <StructureSection load='' size='300' side='right' caption='Morph showing the transition of SARS-CoV-2 spike protein from its closed to open conformation induced by proteolytic cleavage, details at [[SARS-CoV-2_spike_protein_priming_by_furin ]]' scene='85/857141/Morph_alpha_carbons_only/4'> |
| - | [[Image:COVID-19.jpeg|left|200px|thumb|COVID-19 (CDC/Eckert & Higgins)]]
| + | |
| - | A novel coronavirus was identified as the cause of an outbreak of respiratory illness first detected in Wuhan, China in 2019. The illness caused by this virus has been named coronavirus disease 2019 (COVID-19). The illustration (left) shows the overall structure of the COVID-19 virus. Note the spikes that adorn the outer surface, colored <span style="color:red">red</span>, which impart the look of a '''corona''' surrounding the virus. A close up view of these spikes can be seen on the right, from studies by McLellan lab<ref name="McLellan">PMID:32075877</ref>. An animation (by [http://elarasystems.com Elara Systems)] shows how [http://www.dropbox.com/s/24e5f5wdh64f5fv/ES_NL_CoronaVirus_Final.mp4?dl=0 COVID-19 interacts with its host], via these spikes, thus permitting its genome to enter the host (human) cell<ref name="McLellan" />.
| + | |
| - | <br>
| + | |
| - | Photo credit (to the left): Alissa Eckert, MS; Dan Higgins, MAMS
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | == Potential treatments and ways to minimize exposure to COVID-19 ==
| + | |
| - | * A USA, French UK study '''identified 69 drugs to test against the coronavirus'''<ref> Gordon, et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing: bioRxiv (online) 2020[http://doi.org/10.1101/2020.03.22.002386 http://doi.org/10.1101/2020.03.22.002386]</ref>. As reported in the [http://www.nytimes.com/2020/03/22/science/coronavirus-drugs-chloroquine.html?action=click&module=Top%20Stories&pgtype=Homepage New York Times] (23-Mar-2020) "The researchers sought drugs that also latch onto the human proteins that the coronavirus seems to need to enter and replicate in human cells."
| + | |
| - | * [http://youtu.be/s2EVlqql_f8 Fighting Coronavirus with Soap] from PDB-101.
| + | |
| | | | |
| - | * A French study<ref>Gautret, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open- label non-randomized clinical trial: Intl J Antimcrob Agents (in press) 2020 [http://dx.doi.org/10.1016/j.ijantimicag.2020.105949]</ref> showed, despite its small sample size (20 patients treated), that '''hydroxychloroquine treatment is significantly associated with viral load reduction/disappearance in COVID-19 patients''' and its effect is reinforced by azithromycin.
| + | [[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|left|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] |
| | + | <span style="font-size:100%"> |
| | + | A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named '''''COVID-19'''''<ref>[http://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Naming the coronavirus disease (COVID-19) and the virus that causes it]</ref> ('''CO'''rona '''VI'''rus '''D'''isease-20'''19'''). |
| | | | |
| - | == Movies helping to explain COVID-19 ==
| + | There’s a new [http://go.usa.gov/xw9MV NCBI site] on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic. |
| - | * A [http://player.vimeo.com/video/395937110?html5=1&title=1&byline=0&portrait=0&autoplay=0 movie of the 3D model] of the surface of COVID-19 by [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers Fusion Animation].
| + | |
| | | | |
| - | * WHO (Feb 11, 2020) [http://youtu.be/I-Yd-_XIWJg Outbreak COVID-19 explained through 3D Medical Animation].
| + | A video (by [http://elarasystems.com Elara)] shows how the virus [http://youtu.be/hwVl_-lnoys '''interacts with its human (host) cells'''], via its ''spike'' proteins<ref name="Pillay">PMID:32376714</ref><ref name="McLellan">PMID:32075877</ref><ref name="Veesler">PMID:32155444</ref><ref name="wrobel">PMID: 32647346</ref><ref name="thomas">PMID:12360192</ref><ref name="wang-qi">PMID:32275855</ref>, thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at '''[[SARS-CoV-2_protein_S_activation_by_furin]]'''. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at '''[[SARS-CoV-2 spike protein fusion transformation]]'''. |
| | + | </span> |
| | | | |
| - | * Bill Gates Ted Talk (April 3, 2015) on [http://www.youtube.com/watch?v=6Af6b_wyiwI We're Not Ready for the Next Epidemic]. | |
| | | | |
| | == Useful sites on COVID-19 == | | == Useful sites on COVID-19 == |
| - | * [http://deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19 Computational predictions of 3D protein structures associated with COVID-19] based on the [http://deepmind.com/blog/article/AlphaFold-Using-AI-for-scientific-discovery AI AlphaFold system]. Coordinates of the 3D structures can be download [http://storage.googleapis.com/deepmind-com-v3-datasets/alphafold-covid19/structures_4_3_2020.zip here as a '''zip''' file]. | + | * A plot of the Biweekly change in confirmed COVID-19 cases throughout the world |
| | + | <imagemap> |
| | + | Image:Biweekly change in confirmed COVID-19 cases.jpg|500px |
| | + | default [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World site with the most recent data] |
| | + | </imagemap> |
| | + | Clicking the above image goes to the [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World in Data] site with the most recent data. |
| | | | |
| - | * Up to date statistics on [http://www.worldometers.info/coronavirus/coronavirus-cases/ Coronavirus cases world-wide at worldometer] | + | * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ the '''CDC''']. |
| | | | |
| - | * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ CDC]. | + | * A series of fascinating '''[[COVID-19 Videos]]'''. |
| - | * A computer game, [http://youtu.be/gGvlNo3nMfw Folding@home], developed at the [http://www.ipd.uw.edu Inst for Protein Design] (U Washington), uses crowdsourcing to try to find new lead compound that might become drugs to treat COVID-19.
| + | |
| | | | |
| - | * [http://crowdfightcovid19.org Crowdfight COVID-19] - A scientific crowdsourcing initiative to put all available resources at the service of the fight against COVID-19 | + | * '''[[Potential treatments for COVID-19]]'''. |
| | | | |
| - | == Recent published papers ==
| + | * '''[[SARS-CoV-2 virus proteins]]''' - Status of all 3D models of SARS-CoV-2 Virus protein structures. |
| - | * 3D Structure of RNA-dependent RNA polymerase from COVID-19, a '''major antiviral drug target''' from the Rao lab in Beijing<ref> Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020[https://doi.org/10.1101/2020.03.16.993386]</ref>. | + | |
| | | | |
| - | * Crystal structure of the Mpro from COVID-19 and '''discovery of inhibitors''' in a study by scientists from Shanghai & Beijing <ref> Jin, et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors: bioRxiv (online) 2020[http://doi.org/10.1101/2020.02.26.964882]</ref>. | + | * '''[[COVID-19 AlphaFold2 Models]]''' that have been of structures of SARS CoV-2 proteins whose structure have not yet been experimentally determined. |
| | | | |
| - | * '''Coronavirus Evolved Naturally''', and ‘Is '''Not''' a Laboratory Construct,’ in a study in Nature Med by Anderson and colleagues <ref>Andersen, et al. The proximal origin of SARS-CoV-2: Nature Med (in press) 2020[http://dx.doi.org/10.1038/s41591-020-0820-9]</ref>. | + | * '''[[3D structural studies on SARS-CoV-2]]''' - Pointers to research on 3D structural studies on SARS-CoV-2 Virus proteins. |
| | | | |
| - | * A study by Zhou & colleagues in ''Science'' on the structural basis for the recognition of the SARS-CoV-2 (COVID-19) by full-length human ACE2 '''gives insights to the molecular basis for coronavirus recognition and infection'''<ref>PMID:32132184</ref>. | |
| | | | |
| - | * '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />. | |
| | | | |
| - | * '''Scientists are endeavoring to find antivirals specific to the virus'''. Several drugs such as chloroquine, arbidol, remdesivir, and favipiravir are currently undergoing clinical studies to test their efficacy and safety in the treatment of COVID-19 in China, with some promising results summarized.<ref>PMID:32147628</ref>.
| + | == References == |
| | + | <references/> |
| | </StructureSection> | | </StructureSection> |
| | + | |
| | + | [[Category: Coronavirus]] |
| | + | [[Category: Covid-19]] |
| | + | |
| | == References == | | == References == |
| | <references/> | | <references/> |
|
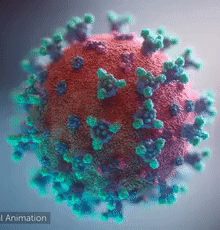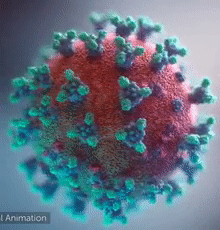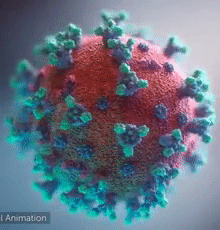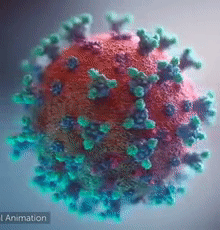 SARS-CoV-2 virus. The spikes, that adorn the virus surface, impart a corona like appearance (Fusion Animation).
A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named COVID-19[1] (COrona VIrus Disease-2019).
There’s a new NCBI site on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic.
A video (by Elara) shows how the virus interacts with its human (host) cells, via its spike proteins[2][3][4][5][6][7], thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at SARS-CoV-2_protein_S_activation_by_furin. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at SARS-CoV-2 spike protein fusion transformation.
Useful sites on COVID-19
- A plot of the Biweekly change in confirmed COVID-19 cases throughout the world
Clicking the above image goes to the Our World in Data site with the most recent data.
- COVID-19 AlphaFold2 Models that have been of structures of SARS CoV-2 proteins whose structure have not yet been experimentally determined.
References
- ↑ Naming the coronavirus disease (COVID-19) and the virus that causes it
- ↑ Pillay TS. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J Clin Pathol. 2020 Jul;73(7):366-369. doi: 10.1136/jclinpath-2020-206658. Epub, 2020 May 6. PMID:32376714 doi:http://dx.doi.org/10.1136/jclinpath-2020-206658
- ↑ Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Feb 19. pii: science.abb2507. doi: 10.1126/science.abb2507. PMID:32075877 doi:http://dx.doi.org/10.1126/science.abb2507
- ↑ Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 Mar 6. pii: S0092-8674(20)30262-2. doi: 10.1016/j.cell.2020.02.058. PMID:32155444 doi:http://dx.doi.org/10.1016/j.cell.2020.02.058
- ↑ Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020 Jul 9. pii: 10.1038/s41594-020-0468-7. doi:, 10.1038/s41594-020-0468-7. PMID:32647346 doi:http://dx.doi.org/10.1038/s41594-020-0468-7
- ↑ Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002 Oct;3(10):753-66. doi: 10.1038/nrm934. PMID:12360192 doi:http://dx.doi.org/10.1038/nrm934
- ↑ Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020 Apr 7. pii: S0092-8674(20)30338-X. doi: 10.1016/j.cell.2020.03.045. PMID:32275855 doi:http://dx.doi.org/10.1016/j.cell.2020.03.045
|



