Coronavirus Disease 2019 (COVID-19)
From Proteopedia
(Difference between revisions)
| (426 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='300' side='right' caption='Morph showing the transition of SARS-CoV-2 spike protein from its closed to open conformation induced by proteolytic cleavage, details at [[SARS-CoV-2_spike_protein_priming_by_furin ]]' scene='85/857141/Morph_alpha_carbons_only/4'> | |
| - | <StructureSection load=' | + | |
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
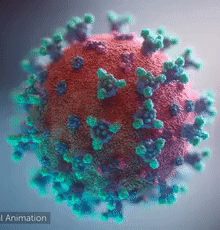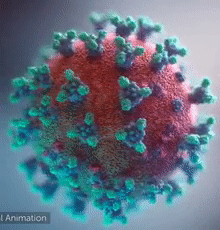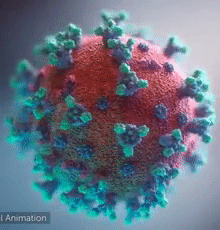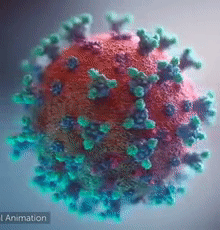
| - | + | [[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|left|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] | |
| + | <span style="font-size:100%"> | ||
| + | A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named '''''COVID-19'''''<ref>[http://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Naming the coronavirus disease (COVID-19) and the virus that causes it]</ref> ('''CO'''rona '''VI'''rus '''D'''isease-20'''19'''). | ||
| - | + | There’s a new [http://go.usa.gov/xw9MV NCBI site] on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | A video (by [http://elarasystems.com Elara)] shows how the virus [http://youtu.be/hwVl_-lnoys '''interacts with its human (host) cells'''], via its ''spike'' proteins<ref name="Pillay">PMID:32376714</ref><ref name="McLellan">PMID:32075877</ref><ref name="Veesler">PMID:32155444</ref><ref name="wrobel">PMID: 32647346</ref><ref name="thomas">PMID:12360192</ref><ref name="wang-qi">PMID:32275855</ref>, thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at '''[[SARS-CoV-2_protein_S_activation_by_furin]]'''. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at '''[[SARS-CoV-2 spike protein fusion transformation]]'''. | |
| + | </span> | ||
| - | * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ CDC]. | ||
| - | * A computer game, developed at the [http://www.ipd.uw.edu Inst for Protein Design] (U Washington), uses crowdsourcing to try to find new lead compound that might become drugs to treat COVID-19. | ||
| - | <html5media height="181" width="322" >https://www.youtube.com/watch?v=gGvlNo3nMfw</html5media> | ||
| - | * [ | + | == Useful sites on COVID-19 == |
| + | * A plot of the Biweekly change in confirmed COVID-19 cases throughout the world | ||
| + | <imagemap> | ||
| + | Image:Biweekly change in confirmed COVID-19 cases.jpg|500px | ||
| + | default [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World site with the most recent data] | ||
| + | </imagemap> | ||
| + | Clicking the above image goes to the [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World in Data] site with the most recent data. | ||
| - | + | * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ the '''CDC''']. | |
| - | * | + | |
| - | * | + | * A series of fascinating '''[[COVID-19 Videos]]'''. |
| - | * | + | * '''[[Potential treatments for COVID-19]]'''. |
| - | * | + | * '''[[SARS-CoV-2 virus proteins]]''' - Status of all 3D models of SARS-CoV-2 Virus protein structures. |
| - | * ''' | + | * '''[[COVID-19 AlphaFold2 Models]]''' that have been of structures of SARS CoV-2 proteins whose structure have not yet been experimentally determined. |
| + | |||
| + | * '''[[3D structural studies on SARS-CoV-2]]''' - Pointers to research on 3D structural studies on SARS-CoV-2 Virus proteins. | ||
| - | * A study by Zhou & colleagues in ''Science'' on the structural basis for the recognition of the SARS-CoV-2 (COVID-19) by full-length human ACE2 '''gives insights to the molecular basis for coronavirus recognition and infection'''<ref>PMID:32132184</ref>. | ||
| - | * '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />, PDB-ID [[6vsb]]. | ||
| - | * '''Scientists are endeavoring to find antivirals specific to the virus'''. Several drugs such as chloroquine, arbidol, remdesivir, and favipiravir are currently undergoing clinical studies to test their efficacy and safety in the treatment of COVID-19 in China, with some promising results summarized.<ref>PMID:32147628</ref>. | ||
== References == | == References == | ||
<references/> | <references/> | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | [[Category: Coronavirus]] | ||
| + | [[Category: Covid-19]] | ||
| + | |||
| + | == References == | ||
| + | <references/> | ||
Current revision
| |||||||||||
References
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Jaime Prilusky, Eric Martz, Jurgen Bosch, David Sehnal, Gianluca Santoni