Coronavirus Disease 2019 (COVID-19)
From Proteopedia
(Difference between revisions)
| (157 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | < | + | <StructureSection load='' size='300' side='right' caption='Morph showing the transition of SARS-CoV-2 spike protein from its closed to open conformation induced by proteolytic cleavage, details at [[SARS-CoV-2_spike_protein_priming_by_furin ]]' scene='85/857141/Morph_alpha_carbons_only/4'> |
| + | |||
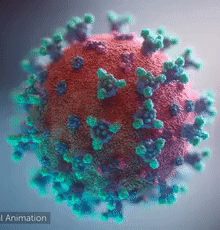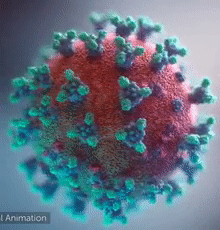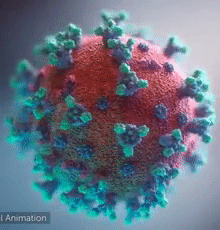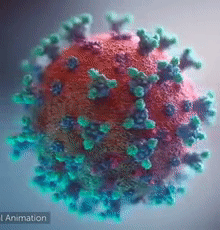
[[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|left|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] | [[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|left|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] | ||
<span style="font-size:100%"> | <span style="font-size:100%"> | ||
| - | A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named '''''COVID-19'''''<ref> | + | A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named '''''COVID-19'''''<ref>[http://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Naming the coronavirus disease (COVID-19) and the virus that causes it]</ref> ('''CO'''rona '''VI'''rus '''D'''isease-20'''19'''). |
| - | + | There’s a new [http://go.usa.gov/xw9MV NCBI site] on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic. | |
| - | + | A video (by [http://elarasystems.com Elara)] shows how the virus [http://youtu.be/hwVl_-lnoys '''interacts with its human (host) cells'''], via its ''spike'' proteins<ref name="Pillay">PMID:32376714</ref><ref name="McLellan">PMID:32075877</ref><ref name="Veesler">PMID:32155444</ref><ref name="wrobel">PMID: 32647346</ref><ref name="thomas">PMID:12360192</ref><ref name="wang-qi">PMID:32275855</ref>, thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at '''[[SARS-CoV-2_protein_S_activation_by_furin]]'''. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at '''[[SARS-CoV-2 spike protein fusion transformation]]'''. | |
</span> | </span> | ||
| - | |||
| - | |||
| - | == Potential treatments for COVID-19 == | ||
| - | * 04-April-2020 Conserved domain analysis, homology modeling, and molecular docking study suggests that SARS-CoV-2 reduces capacity of hemoglobin to carry oxygen, potentiating respiratory syndrome <ref>COVID-19 Disease ORF8 and Surface Glycoprotein Inhibit Heme Metabolism by Binding to Porphyrin [https://chemrxiv.org/articles/COVID-19_Disease_ORF8_and_Surface_Glycoprotein_Inhibit_Heme_Metabolism_by_Binding_to_Porphyrin/11938173/4]</ref> | ||
| - | * [http://economictimes.indiatimes.com/news/international/world-news/oxford-university-begins-enrolling-over-500-volunteers-for-coronavirus-vaccine-trial/articleshow/74864754.cms?utm_source=contentofinterest&utm_medium=text&utm_campaign=cppst Oxford Univ begins enrolling over 500 volunteers for a '''coronavirus vaccine trial'''] also see more at [http://www.jenner.ac.uk '''The Jenner Inst''']. | ||
| - | |||
| - | * An int'l group of scientists (UK, Israel & USA) are trying to combat COVID-19 via a massive crystal-based 3D fragment screen. They determined the 3D structures of [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem/Downloads.html '''over 90 fragments with 66 in the active site''']. All details are available [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem.html '''at the Diamond Light Source''']. They welcome contributions in many different forms, see [http://covid.postera.ai/covid '''PostEra''']. | ||
| - | |||
| - | * 31-Mar-2020 ''NY Times'' [http://www.nytimes.com/2020/03/31/health/cdc-masks-coronavirus.html C.D.C. Weighs Advising Everyone to Wear a Mask]. | ||
| - | |||
| - | * 27-Mar-2020 ''Science'' - Not wearing masks to protect against coronavirus is a '''big mistake''', Dr. George Gao, Director General of the Chinese CDC says in [http://www.sciencemag.org/news/2020/03/not-wearing-masks-protect-against-coronavirus-big-mistake-top-chinese-scientist-says ''Science'']. | ||
| - | |||
| - | * 23-Mar-2020 A USA, French & UK study '''identified 69 drugs to test against the coronavirus'''<ref> Gordon, et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.22.002386 http://doi.org/10.1101/2020.03.22.002386]</ref>. As reported in the [http://www.nytimes.com/2020/03/22/science/coronavirus-drugs-chloroquine.html?action=click&module=Top%20Stories&pgtype=Homepage ''NY Times'']. | ||
| - | |||
| - | * 20-Mar-2020 - Analysis by Abby Olena in ''The Scientist'' on [http://www.the-scientist.com/news-opinion/remdesivir-works-great-against-coronaviruses-in-the-lab-67298?utm_campaign=TS_OTC_2020&utm_source=hs_email&utm_medium=email&utm_content=85306117&_hsenc=p2ANqtz--t49MmUS_nL_8CL8wosEbymFBDq_4smPhI1pSJmSKWA0bURCpjoMb1eYxIVv30SDzsuiNd9PRGTPC30x-gelTGjK-9r9wPWaMjEZIqjaz5Y0tgW5w&_hsmi=85306117 '''Remdesivir Works Against Coronaviruses in the Lab'''] | ||
| - | |||
| - | * 20-Mar-2020 - Analysis by Chris Baraniuk in ''The Scientist'' on [http://www.the-scientist.com/news-opinion/is-hype-over-chloroquine-as-a-potential-covid-19-therapy-justified--67301?utm_campaign=TS_OTC_2020&utm_source=hs_email&utm_medium=email&utm_content=85306117&_hsenc=p2ANqtz--t49MmUS_nL_8CL8wosEbymFBDq_4smPhI1pSJmSKWA0bURCpjoMb1eYxIVv30SDzsuiNd9PRGTPC30x-gelTGjK-9r9wPWaMjEZIqjaz5Y0tgW5w&_hsmi=85306117 '''Chloroquine for COVID-19: Cutting Through the Hype'''] | ||
| - | |||
| - | * A French study<ref> Gautret, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open- label non-randomized clinical trial: Intl J Antimcrob Agents (in press) 2020 [http://dx.doi.org/10.1016/j.ijantimicag.2020.105949 http://dx.doi.org/10.1016/j.ijantimicag.2020.105949]</ref> showed despite a small sample size (20 patients treated), that '''hydroxychloroquine is significantly associated with viral load reduction & disappearance in COVID-19 patients''' & its effect is reinforced by azithromycin. | ||
| - | |||
| - | == Videos helping to explain COVID-19 == | ||
| - | <html5media height="225" width="400">http://www.youtube.com/watch?v=HhNo_IOPOtU</html5media><br> | ||
| - | The Czech Republic took the uncommon step of making '''wearing of masks mandatory''' in public spaces, prompting a [http://www.sciencemag.org/news/2020/03/would-everyone-wearing-face-masks-help-us-slow-pandemic '''grassroots effort to make masks'''], with '''wonderful''' results<br>. | ||
| - | <br> | ||
| - | <html5media height="160" width="400">https://w.soundcloud.com/player/?url=https%3A//api.soundcloud.com/tracks/775538998&color=%23ff5500&auto_play=false&hide_related=false&show_comments=true&show_user=true&show_reposts=false&show_teaser=true</html5media><br> | ||
| - | Scientists have turned the coronavirus spike protein structure into '''music(!!)''', as reported in [http://www.sciencemag.org/news/2020/04/scientists-have-turned-structure-coronavirus-music?fbclid=IwAR0-kIT3mVjRHe18Y7wf3KrqaBIvIk9JGG9KuJsbA0zN6gsov0Np0p_jO9A# ''Science''] by [http://soundcloud.com/user-275864738/viral-counterpoint-of-the-coronavirus-spike-protein-2019-ncov Soundcloud].<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">http://www.youtube.com/watch?v=hm7BmIJrLZE</html5media><br> | ||
| - | Japan researchers show how '''Coronavirus spreads through micro droplets'''. These tiny droplets containing the virus can stay in the air for extended periods.<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">http://www.youtube.com/watch?v=qzARpgx8cvE</html5media><br> | ||
| - | Speaking face-to-face is exchanging saliva, so '''stop speaking face-to-face and stay healthy''' by [http://stoptheviruscovid19.github.io '''stoptheviruscovid'''].<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">http://www.youtube.com/watch?v=Kas0tIxDvrg</html5media><br> | ||
| - | Modeling an epidemic (6-Mar-2020) '''''We're not ready for the next epidemic''''' by [http://3b1b.co/covid-thanks '''3blue1brown'''].<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">https://www.youtube.com/watch?v=s2EVlqql_f8</html5media><br> | ||
| - | '''Fighting Coronavirus with Soap''' by [http://pdb101.rcsb.org/ '''PDB-101'''].<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">https://www.youtube.com/watch?v=I0TmBsHaGmI</html5media><br> | ||
| - | '''Animation''' of '''3D structure/function of Coronavirus-CoV-2 proteins''' by [http://www.biolution.net '''biolution GmBH'''].<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">https://www.youtube.com/watch?v=I-Yd-_XIWJg</html5media><br> | ||
| - | '''COVID-19 outbreak shown via 3D animation (11-Feb 11-2020)''' by [http://www.scientificanimations.com '''Sci Animations'''].<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">http://www.youtube.com/watch?v=6Af6b_wyiwI</html5media><br> | ||
| - | '''Bill Gates Ted Talk (3-Apr-2015) ''We're not ready for the next epidemic''''' by [http://www.ted.com/talks '''Ted Talks''']. | ||
| - | <br> | ||
| - | <html5media height="225" width="400">http://www.youtube.com/watch?v=uSDC5L7qYUc</html5media><br> | ||
| - | '''George W. Bush''' urged us to '''prepare for future pandemics in 2005''', at the NIH.<br> | ||
| - | <br> | ||
| - | <html5media height="225" width="400">https://www.youtube.com/watch?v=B00tJnbktVo&feature=youtu.be&t=210</html5media><br> | ||
| - | '''Jürgen Bosch Presentation (31-Mar-2020) ''Coronavirus: How it Ticks and What we are Doing to Stop it''''' by [http://www.linkedin.com/in/jubosch '''Jürgen Bosch''']. | ||
== Useful sites on COVID-19 == | == Useful sites on COVID-19 == | ||
| - | * | + | * A plot of the Biweekly change in confirmed COVID-19 cases throughout the world |
| - | + | <imagemap> | |
| - | + | Image:Biweekly change in confirmed COVID-19 cases.jpg|500px | |
| + | default [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World site with the most recent data] | ||
| + | </imagemap> | ||
| + | Clicking the above image goes to the [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World in Data] site with the most recent data. | ||
* A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ the '''CDC''']. | * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ the '''CDC''']. | ||
| - | * ''' | + | * A series of fascinating '''[[COVID-19 Videos]]'''. |
| - | * ''' | + | * '''[[Potential treatments for COVID-19]]'''. |
| - | * | + | * '''[[SARS-CoV-2 virus proteins]]''' - Status of all 3D models of SARS-CoV-2 Virus protein structures. |
| - | + | ||
| - | + | * '''[[COVID-19 AlphaFold2 Models]]''' that have been of structures of SARS CoV-2 proteins whose structure have not yet been experimentally determined. | |
| - | + | ||
| - | + | ||
| - | * [[SARS-CoV-2 | + | * '''[[3D structural studies on SARS-CoV-2]]''' - Pointers to research on 3D structural studies on SARS-CoV-2 Virus proteins. |
| - | == 3D structural studies on SARS-CoV-2 virus== | ||
| - | |||
| - | * A team of UK & Israeli scientists determined the 3D structures of [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem/Downloads.html '''over 90 fragments, 66 of which are in the active site'']. All the experimental details and results are available [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem.html '''online at the Diamond Light Source''']. | ||
| - | |||
| - | * A team of Chinese scientists determined, by Cryo-EM, the''' coronavirus spike receptor-binding domain complexed with its receptor ACE2 PDB-ID''' [http://www.rcsb.org/structure/6LZG 6LZG]. (To be published). | ||
| - | |||
| - | * A team of US and Chinese scientists determined the crystal structure of '''2019-nCoV spike receptor-binding domain bound with ACE2''' [http://www.rcsb.org/structure/6M0J 6M0J] (To be published). | ||
| - | |||
| - | * A team of US scientists determined, by Cryo-EM, the '''structure of the SARS-CoV-2 spike glycoprotein (open & closed states)''' <ref>PMID:32155444</ref>, PDB-ID [http://www.rcsb.org/structure/6VXX 6VXX] & [http://www.rcsb.org/structure/6VYB 6VYB] | ||
| - | |||
| - | * Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 [http://www.rcsb.org/structure/6W41 6W41] (To be published). | ||
| - | |||
| - | * Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2 [http://www.rcsb.org/structure/6W61 6W61] (To be published). | ||
| - | |||
| - | * Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP [http://www.rcsb.org/structure/6W6Y 6W6Y] (To be published). | ||
| - | |||
| - | * Structure of NSP10 - NSP16 Complex from SARS-CoV-2 [http://www.rcsb.org/structure/6W75 6W75] (To be published). | ||
| - | |||
| - | * Crystal structure of SARS-CoV-2 nucleocapsid protein N-terminal RNA binding domain [http://www.rcsb.org/structure/6M3M 6M3M] (To be published). | ||
| - | |||
| - | * Crystal structure of Nsp9 RNA binding protein of SARS CoV-2 [http://www.rcsb.org/structure/6W4B 6W4B] (To be published). | ||
| - | |||
| - | * Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 [http://www.rcsb.org/structure/6W4H 6W4H] (To be published). | ||
| - | |||
| - | * Cryo-EM structure of the 2019-nCoV RBD/ACE2-B0AT1 complex<ref>PMID:32132184</ref> [http://www.rcsb.org/structure/6M17 6M17]. | ||
| - | |||
| - | * Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 [http://www.rcsb.org/structure/6VYO 6VYO] (To be published). | ||
| - | |||
| - | * Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate [http://www.rcsb.org/structure/6W01 6W01] (To be published). | ||
| - | |||
| - | * Crystal structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose [http://www.rcsb.org/structure/6W02 6W02] (To be published). | ||
| - | |||
| - | * Crystal structure of 2019-nCoV chimeric receptor-binding domain complexed with its receptor human ACE2 [http://www.rcsb.org/structure/6VW1 6VW1] (To be published). | ||
| - | |||
| - | * Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 [http://www.rcsb.org/structure/6VWW 6VWW] (To be published). | ||
| - | |||
| - | * Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 [http://www.rcsb.org/structure/6VXS 6VXS] (To be published). | ||
| - | |||
| - | * Crystal structure of the 2019-nCoV HR2 Domain [http://www.rcsb.org/structure/6LVN 6LVN] (To be published). | ||
| - | |||
| - | * Crystal structure of post fusion core of 2019-nCoV S2 subunit [http://www.rcsb.org/structure/6LXT 6LXT] (To be published). | ||
| - | |||
| - | * Crystal structure of '''SARS-CoV-2 main protease''' provides a basis for design of improved α-ketoamide inhibitors, from the Hilgenfeld lab<ref>PMID:32198291</ref>, Apo Struture: PDB-ID [http://www.rcsb.org/structure/6y2e 6Y2E], and complexes with inhibitors: PDB-ID [http://www.rcsb.org/structure/6y2f 6Y2F] and [http://www.rcsb.org/structure/6y2g 6Y2G]. | ||
| - | |||
| - | * 3D Structure of '''RNA-dependent RNA polymerase from COVID-19''', a '''major antiviral drug target''' from the Rao lab in Beijing<ref> Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020 [https://doi.org/10.1101/2020.03.16.993386 https://doi.org/10.1101/2020.03.16.993386]</ref>. | ||
| - | |||
| - | * Crystal structure of the '''Mpro from COVID-19''' and '''discovery of inhibitors''' in a study by scientists from Shanghai & Beijing <ref> Jin, et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.02.26.964882 http://doi.org/10.1101/2020.02.26.964882]</ref>, PDB-ID [[2h2z]]. | ||
| - | |||
| - | * Crystal structure of '''Nsp15 endoribonuclease NendoU from SARS-CoV-2''' in a study by scientists from USA<ref> Kim, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.02.968388 http://doi.org/10.1101/2020.03.02.968388]</ref>, PDB-ID [[6w01]]. | ||
| - | |||
| - | * A study by Zhou & colleagues on the structural basis for the '''recognition of the SARS-CoV-2 (COVID-19) by full-length human ACE2''' gives insights to the molecular basis for coronavirus recognition and infection<ref>PMID:32132184</ref>. | ||
| - | |||
| - | * '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />, PDB-ID [[6vsb]]. | ||
| - | |||
| - | * [http://github.com/thorn-lab/coronavirus_structural_task_force Public resource for the structures from beta-coronavirus with a focus on SARS-CoV and SARS-CoV-2] | ||
| + | == References == | ||
| + | <references/> | ||
| + | </StructureSection> | ||
| + | [[Category: Coronavirus]] | ||
| + | [[Category: Covid-19]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | </SX> | ||
| - | [[Category: Coronavirus]] | ||
| - | [[Category: Covid-19]] | ||
Current revision
| |||||||||||
References
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Jaime Prilusky, Eric Martz, Jurgen Bosch, David Sehnal, Gianluca Santoni