This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 897
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
== Overview == | == Overview == | ||
| - | Bromodomain and PHD finger-containing protein family is the group of proteins found in higher eukaryotes. The members have similar structural features and believed to be an essential part of interaction on H3K36me3.<ref>Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010 Sep 17;142(6):967-80.</ref> The PWWP motif, a part of BRPF proteins, belongs to the Royal family, which consists of Tudor, chromodomain, MBT(Malignant Brain Tumor), and PWWP domains. The PWWP domain was first identified as a structural motif includes 100 to 130 amino acids in WHSC1 protein research. The researchers named after its very conservative sequence, Pro-Trp-Trp-Pro located at the β2 strand of protein.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> This domain is mainly related to the recognition of methylated histone tail lysine and epigenetic regulation of genes. Unlike BRPF1 and 2, BRPF3 is less studied by the researchers. Therefore many of its function remains unknown. | + | Bromodomain and PHD finger-containing protein family is the group of proteins found in higher eukaryotes. The members have similar structural features and believed to be an essential part of interaction on H3K36me3.<ref>Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010 Sep 17;142(6):967-80.</ref> The PWWP motif, a part of BRPF proteins, belongs to the Royal family, which consists of Tudor, chromodomain, MBT(Malignant Brain Tumor), and PWWP domains. The PWWP domain was first identified as a structural motif includes 100 to 130 amino acids in WHSC1 protein research. The researchers named after its very conservative sequence, Pro-Trp-Trp-Pro located at the β2 strand of protein.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> This domain is mainly related to the recognition of specific methylated histone tail lysine and epigenetic regulation of genes. Unlike BRPF1 and 2, BRPF3 is less studied by the researchers. Therefore many of its function remains unknown. |
== Structure == | == Structure == | ||
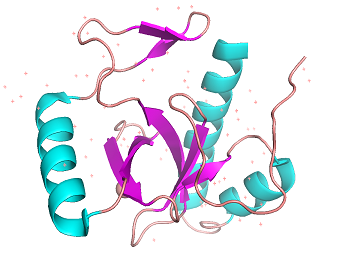
| - | The main characteristic of PWWP domains is the two distinctive substructural motifs: β-barrel at the N-terminal region and helixes at C-terminal region.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> 3PFS has a β-barrel composed of 5 β-strands, which is a very conservative feature of PWWP domains. The helixes at the C-terminal region are not very conservative; 3pfs has 3 helixes which are little more than many of its relatives. The Pro-Trp-Trp-Pro motif, which is the most conservative, is a little different from its name, just like its closely-related proteins. The PWWP domains in the BRPF family are composed of Tyr-Pro-Ser-Tyr and may suggest they would have a similar affinity. The stability of domain comes from both inter-substructure and intra-substructure interactions including hydrogen bonds and polar interactions. One unique feature of the PWWP motif is that the first position affects the stability and aggregation of the protein. The proline gives more stability and oligomerization to the protein, compared to the alanine at the same position.<ref>Hung YL, Lee HJ, Jiang I, Lin SC, Lo WC, Lin YJ, Sue SC. The first residue of the PWWP motif modulates HATH domain binding, Stability, and Protein–Protein Interaction. Biochemistry. 2015 Jul 7;54(26):4063-74.</ref> | + | The main characteristic of PWWP domains is the two distinctive substructural motifs: β-barrel at the N-terminal region and helixes at C-terminal region.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> 3PFS has a β-barrel composed of 5 β-strands, which is a very conservative feature of PWWP domains. The helixes at the C-terminal region are not very conservative; 3pfs has 3 helixes which are little more than many of its relatives. The Pro-Trp-Trp-Pro motif, which is the most conservative, is a little different from its name, just like its closely-related proteins. The PWWP domains in the BRPF family are composed of Tyr-Pro-Ser-Tyr and may suggest they would have a similar affinity. The stability of domain comes from both inter-substructure and intra-substructure interactions including hydrogen bonds and polar interactions. One unique feature of the PWWP motif is that the first position of arrangement affects the stability and aggregation of the protein. The proline gives more stability and oligomerization to the protein, compared to the alanine at the same position.<ref>Hung YL, Lee HJ, Jiang I, Lin SC, Lo WC, Lin YJ, Sue SC. The first residue of the PWWP motif modulates HATH domain binding, Stability, and Protein–Protein Interaction. Biochemistry. 2015 Jul 7;54(26):4063-74.</ref> |
[[Image:3pfs cartoon.png|thumb|center|512 px| '''Figure 1A:''' Cartoon model of 3PFS motif. β-sheets(magenta) and helixes(orange) are shown. generated in PyMOL using PDB: ''3PFS'']] | [[Image:3pfs cartoon.png|thumb|center|512 px| '''Figure 1A:''' Cartoon model of 3PFS motif. β-sheets(magenta) and helixes(orange) are shown. generated in PyMOL using PDB: ''3PFS'']] | ||
[[Image:3pfs alignment.png|thumb|center|512 px| '''Figure 1B:''' The alignment of BRPF1 PWWP domain and BRPF3 PWWP domain. The green box indicates the 'PWWP' motif. generated in Tcoffee using PDB: ''3PFS', '2X35'']] | [[Image:3pfs alignment.png|thumb|center|512 px| '''Figure 1B:''' The alignment of BRPF1 PWWP domain and BRPF3 PWWP domain. The green box indicates the 'PWWP' motif. generated in Tcoffee using PDB: ''3PFS', '2X35'']] | ||
| Line 15: | Line 15: | ||
===Methyl-lysine reader=== | ===Methyl-lysine reader=== | ||
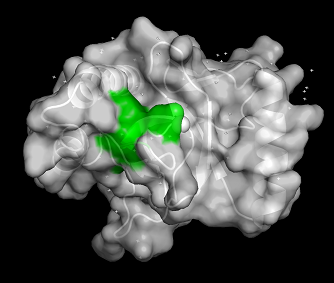
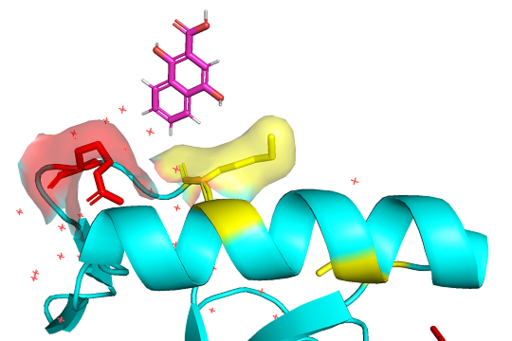
| - | PWWP domains are one of the epigenetic regulators which recognize specific lysine residues that are methylated. Even though there is no detailed research on 3pfs, it may be assumed to have similar function with BRPF1 PWWP domain which is shown to have methylated histone binding activity.<ref>Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Göttgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010 May;17(5):617-9.</ref> With a high-throughput mass spectrometry screening, | + | PWWP domains are one of the epigenetic regulators which recognize specific lysine residues that are methylated. Even though there is no detailed research on 3pfs, it may be assumed to have similar function with BRPF1 PWWP domain which is shown to have methylated histone binding activity.<ref>Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Göttgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010 May;17(5):617-9.</ref> With a high-throughput mass spectrometry screening, 3PFS is also suggested as an essential domain of histone 3 lysine 36 trimethylation binding.<ref>Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010 Sep 17;142(6):967-80.</ref> The aromatic residues on β-strands and Pro-Trp-Trp-Pro motif form hydrophobic cavity, which is the binding site of methylated lysines.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> Since the aromatic cage is a common structure in the Royal family, it can be a common tool for methylated lysine binding.<ref>Qiu Y, Zhang W, Zhao C, Wang Y, Wang W, Zhang J, Zhang Z, Li G, Shi Y, Tu X, Wu J. Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochemical Journal. 2012 Mar 15;442(3):527-38.</ref> |
[[Image:PWWP surface.png|thumb|center|512 px| '''Figure 2:''' Surface of 3PFS with PWWP motif(green) is indicated. generated in PyMOL using PDB: ''3PFS'']] | [[Image:PWWP surface.png|thumb|center|512 px| '''Figure 2:''' Surface of 3PFS with PWWP motif(green) is indicated. generated in PyMOL using PDB: ''3PFS'']] | ||
| Line 25: | Line 25: | ||
=== Evolutionary conservations === | === Evolutionary conservations === | ||
| - | PWWP domain is only found in eukaryotes, from a yeast to a human. The number of PWWP containing protein varies on species; a man has 20 PWWP containing proteins. The loyal family has a common structural characteristic, three conserved β-strands.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> Therefore, PWWP domains are believed to be evolved from a common ancestor which had 3 β-strands. Using the domain-tree approach, the BRPF family has branched out from rest of PWWP domain-containing proteins early and evolved itself.<ref>Alvarez-Venegas R, Avramova Z. Evolution of the PWWP-domain encoding genes in the plant and animal lineages. BMC evolutionary biology. 2012 Dec 1;12(1):101.</ref> As a domain, the PWWP presents in both unicellular and multicellular organisms. However, proteins of unicellular organisms are not found in multicellular creatures. Therefore, the PWWP domain was transmitted as a conserved linear arrangement alone, not with the whole protein.<ref>Alvarez-Venegas R, Avramova Z. Evolution of the PWWP-domain encoding genes in the plant and animal lineages. BMC evolutionary biology. 2012 Dec 1;12(1):101.</ref> | + | PWWP domain is only found in eukaryotes, from a yeast to a human. The number of PWWP containing protein varies on species; a man has 20 PWWP containing proteins. The loyal family has a common structural characteristic, three conserved β-strands.<ref>Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.</ref> Therefore, PWWP domains are believed to be evolved from a common ancestor which had 3 β-strands. Using the domain-tree approach, it is suggested that the BRPF family has branched out from rest of PWWP domain-containing proteins early and evolved itself.<ref>Alvarez-Venegas R, Avramova Z. Evolution of the PWWP-domain encoding genes in the plant and animal lineages. BMC evolutionary biology. 2012 Dec 1;12(1):101.</ref> As a domain, the PWWP presents in both unicellular and multicellular organisms. However, proteins of unicellular organisms are not found in multicellular creatures. Therefore, the PWWP domain was transmitted as a conserved linear arrangement alone, not with the whole protein.<ref>Alvarez-Venegas R, Avramova Z. Evolution of the PWWP-domain encoding genes in the plant and animal lineages. BMC evolutionary biology. 2012 Dec 1;12(1):101.</ref> |
== Therapheutic features == | == Therapheutic features == | ||
Current revision
| This Sandbox is Reserved from Jan 13 through July 31, 2020 for use in the course Protein Structure in Drug Discovery taught by Karen C. Glass at the ACPHS, Colchester, United States. This reservation includes Sandbox Reserved 895 through Sandbox Reserved 901. |
To get started:
More help: Help:Editing |
BRPF3 PWWP binding domain 3PFS
| |||||||||||
References
- ↑ Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010 Sep 17;142(6):967-80.
- ↑ Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.
- ↑ Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.
- ↑ Hung YL, Lee HJ, Jiang I, Lin SC, Lo WC, Lin YJ, Sue SC. The first residue of the PWWP motif modulates HATH domain binding, Stability, and Protein–Protein Interaction. Biochemistry. 2015 Jul 7;54(26):4063-74.
- ↑ Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Göttgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010 May;17(5):617-9.
- ↑ Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010 Sep 17;142(6):967-80.
- ↑ Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.
- ↑ Qiu Y, Zhang W, Zhao C, Wang Y, Wang W, Zhang J, Zhang Z, Li G, Shi Y, Tu X, Wu J. Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochemical Journal. 2012 Mar 15;442(3):527-38.
- ↑ Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nature structural biology. 2002 Mar;9(3):217-24.
- ↑ Rona GB, Eleutherio EC, Pinheiro AS. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophysical reviews. 2016 Mar 1;8(1):63-74.
- ↑ Alvarez-Venegas R, Avramova Z. Evolution of the PWWP-domain encoding genes in the plant and animal lineages. BMC evolutionary biology. 2012 Dec 1;12(1):101.
- ↑ Alvarez-Venegas R, Avramova Z. Evolution of the PWWP-domain encoding genes in the plant and animal lineages. BMC evolutionary biology. 2012 Dec 1;12(1):101.
- ↑ Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Göttgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010 May;17(5):617-9.
- ↑ Bjarkam CR, Corydon TJ, Olsen IM, Pallesen J, Nyegaard M, Fryland T, Mors O, Børglum AD. Further immunohistochemical characterization of BRD1 a new susceptibility gene for schizophrenia, and bipolar affective disorder. Brain Structure and Function. 2009 Dec 1;214(1):37.