This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Sumit Kamat/Sandbox Reserved 901
From Proteopedia
< User:Sumit Kamat(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 20: | Line 20: | ||
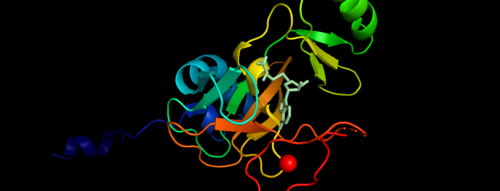
| - | [[Image:Zinc Binding.png|thumb|750px|center|Figure 4. | + | [[Image:Zinc Binding.png|thumb|750px|center|Figure 4. KMT2A SET Domain with the cofactor product S-Adenosylhomocysteine and zinc. <ref> The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. </ref> ]] |
| Line 26: | Line 26: | ||
MLL-1 is a 431-kDa protein to be a structural and functional homolog of the Drosophila trithorax (TRX) protein. Two domains are highly conserved between MLL and TRX consist of a carboxy-terminal SET (Su(var)3-9, enhancer-of-zeste, and trithorax) domain and internal plant homeodomain (PHD) fingers. Both domains are found in many chromatin-associated transcriptional regulators and are thought to function either directly in chromatin modification or as protein-protein interaction surfaces for the recruitment of chromatin-modifying machinery <ref> Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties | MLL-1 is a 431-kDa protein to be a structural and functional homolog of the Drosophila trithorax (TRX) protein. Two domains are highly conserved between MLL and TRX consist of a carboxy-terminal SET (Su(var)3-9, enhancer-of-zeste, and trithorax) domain and internal plant homeodomain (PHD) fingers. Both domains are found in many chromatin-associated transcriptional regulators and are thought to function either directly in chromatin modification or as protein-protein interaction surfaces for the recruitment of chromatin-modifying machinery <ref> Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties | ||
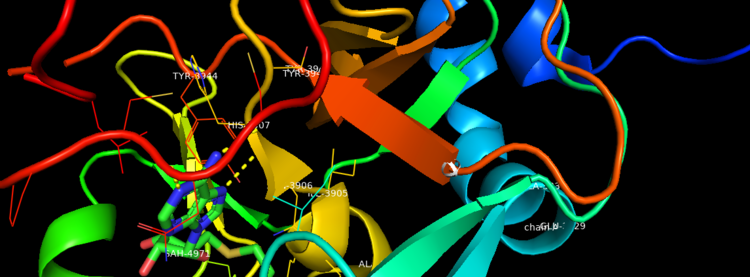
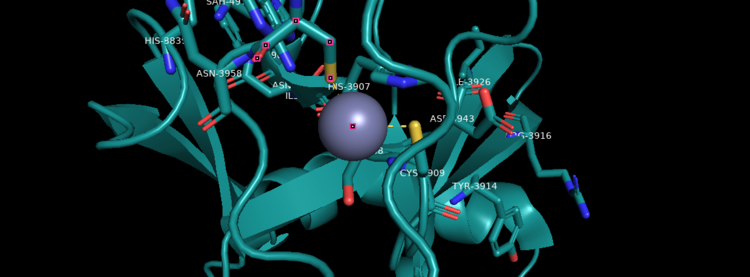
| - | Akihiko Yokoyama, Issay Kitabayashi, Paul M. Ayton, Michael L. Cleary, Misao Ohki https://doi.org/10.1182/blood-2002-04-1015 </ref>.There are no significant changes in the structure of the SET domain of the binary complex with the cofactor binding in the surface pocket. It has been determined at the essential active site residues like Phe3884, Tyr3942, Tyr3944, and Phe3946 and the main chain of the tetrapeptide, residues Cys3882 to Phe3885 have a similar arrangement to other SET domains earlier characterized. In the absence of substrate, the electron density for the side chain of Tyr3942 suggests two orientations, but upon substrate binding it becomes ordered and assumes the rotamer equivalent to other structures <ref> Structural Basis for the Requirement of Additional Factors for MLL1 SET Domain Activity and Recognition of Epigenetic Marks | + | Akihiko Yokoyama, Issay Kitabayashi, Paul M. Ayton, Michael L. Cleary, Misao Ohki https://doi.org/10.1182/blood-2002-04-1015 </ref>. The protein structure consists of 7 helices which comprise of almost 19 percent of the structure, it also has 14 strands of beta sheets which comprise of 26 percent of the structure.There are no significant changes in the structure of the SET domain of the binary complex with the cofactor binding in the surface pocket. It has been determined at the essential active site residues like Phe3884, Tyr3942, Tyr3944, and Phe3946 and the main chain of the tetrapeptide, residues Cys3882 to Phe3885 have a similar arrangement to other SET domains earlier characterized. In the absence of substrate, the electron density for the side chain of Tyr3942 suggests two orientations, but upon substrate binding it becomes ordered and assumes the rotamer equivalent to other structures <ref> Structural Basis for the Requirement of Additional Factors for MLL1 SET Domain Activity and Recognition of Epigenetic Marks |
Stacey M. Southall 2 Poon-Sheng Wong 2 Zain Odho S. Mark Roe Jon R. Wilson ArchiveDOI:https://doi.org/10.1016/j.molcel.2008.12.029 </ref>. | Stacey M. Southall 2 Poon-Sheng Wong 2 Zain Odho S. Mark Roe Jon R. Wilson ArchiveDOI:https://doi.org/10.1016/j.molcel.2008.12.029 </ref>. | ||
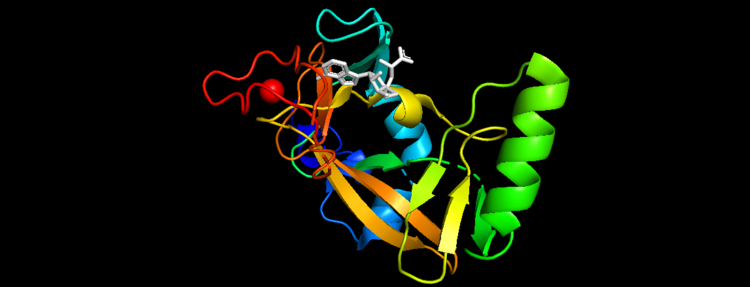
| - | [[Image:2w5y sequence chain final.PNG|thumb|750px|center|Figure | + | [[Image: New secondary.png|thumb|750px|center|Figure 5. Secondary Structure of KMT2A SET Domain with the cofactor product S-Adenosylhomocysteine <ref> The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. </ref> ]] |
| + | |||
| + | |||
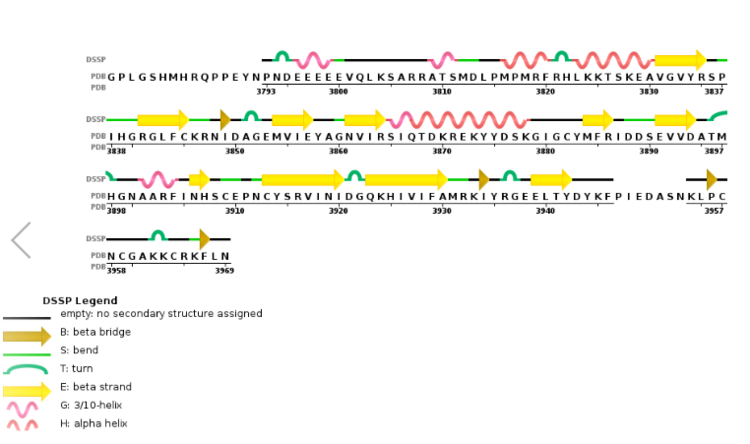
| + | [[Image:2w5y sequence chain final.PNG|thumb|750px|center|Figure 6. Secondary structure of KMT2A SET Domain with the cofactor product S-Adenosylhomocysteine. <ref> PMID: 6667333 </ref> ]] | ||
| Line 37: | Line 40: | ||
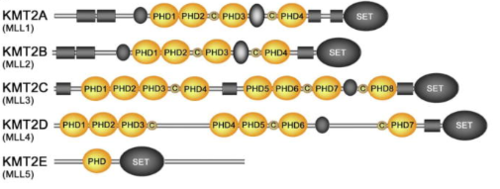
KMT2A alone has over a dozen of binding partners and is cleaved into two pieces, a larger N-terminal fragment, involved in gene repression, and a smaller C-terminal fragment, which is a transcriptional activator. The cleavage, followed by the association of the two fragments, is necessary for KMT2A to be fully active. KMT2A-E can be distinguished through the catalytic Su(var)3–9, Enhancer of Zeste, Trithorax (SET) domain, however the number of PHD fingers found in these proteins differs considerably. Four PHD fingers are present in KMT2A and KMT2B, whereas KMT2C, KMT2D and KMT2E have eight, seven, and one, respectively <ref> Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23(1):186–194. doi:10.1128/mcb.23.1.186-194.2003 </ref>. | KMT2A alone has over a dozen of binding partners and is cleaved into two pieces, a larger N-terminal fragment, involved in gene repression, and a smaller C-terminal fragment, which is a transcriptional activator. The cleavage, followed by the association of the two fragments, is necessary for KMT2A to be fully active. KMT2A-E can be distinguished through the catalytic Su(var)3–9, Enhancer of Zeste, Trithorax (SET) domain, however the number of PHD fingers found in these proteins differs considerably. Four PHD fingers are present in KMT2A and KMT2B, whereas KMT2C, KMT2D and KMT2E have eight, seven, and one, respectively <ref> Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23(1):186–194. doi:10.1128/mcb.23.1.186-194.2003 </ref>. | ||
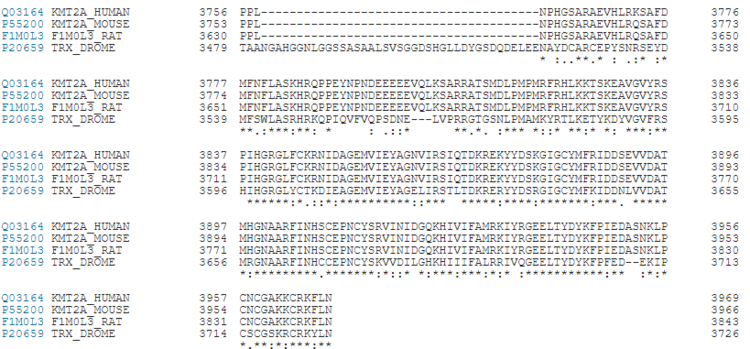
| - | [[Image: Kmt2a conservation hmrf.PNG |thumb|750px|center|Figure | + | [[Image: Kmt2a conservation hmrf.PNG |thumb|750px|center|Figure 7. KMT2A Conservation between Humans, Rats, Mouse and Fruit Flies <ref> UniProt: a worldwide hub of protein knowledge Nucleic Acids Res. 47: D506-515 (2019) </ref> ]] |
| - | [[Image:Conservation final 2.PNG |thumb|500px|center|Figure | + | [[Image:Conservation final 2.PNG |thumb|500px|center|Figure 8.Schematic representation of the KMT2A-E subfamily ]] |
Current revision
Histone-lysine N-methyltransferase 2A KMT2A
| |||||||||||