User:Tania Girao Mangolini/Sandbox 1
From Proteopedia
| (87 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==HORSERADISH PEROXIDASE C1A== | ==HORSERADISH PEROXIDASE C1A== | ||
<Structure load='1h58' size='350' frame='true' align='right' caption='' scene='84/845930/Hrp_c1a/2' /> | <Structure load='1h58' size='350' frame='true' align='right' caption='' scene='84/845930/Hrp_c1a/2' /> | ||
| - | Horseadish ([http://en.wikipedia.org/wiki/Horseradish ''Armoracia rusticana'']) is a plant that belongs to the [http://en.wikipedia.org/wiki/Brassicaceae Brassicaceae] family. The roots of this plant are rich in [http://en.wikipedia.org/wiki/Peroxidase peroxidases], being the HRP C [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4322221/table/Tab1/?report=objectonly isozymes] the most common ones. <ref name="review">Veitch, N.C. Horseadich peroxidase: a modern view of a classic enzyme 65:249-259 (2004). [https://doi.org/10.1016/j.phytochem.2003.10.022 DOI: 10.1016/j.phytochem.2003.10.022]</ref> | + | Horseadish ([http://en.wikipedia.org/wiki/Horseradish ''Armoracia rusticana'']) is a plant that belongs to the [http://en.wikipedia.org/wiki/Brassicaceae Brassicaceae] family. The roots of this plant are rich in class III [http://en.wikipedia.org/wiki/Peroxidase peroxidases]<ref name="ref4">Pandey, Veda & Awasthi, Manika & Singh, Swati & Tiwari, Sameeksha & Dwivedi, Upendra. (2017). A Comprehensive Review on Function and Application of Plant Peroxidases. Biochemistry & Analytical Biochemistry. [https://doi.org/10.4172/2161-1009.1000308 DOI: 10.4172/2161-1009.1000308]</ref>, being the HRP C [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4322221/table/Tab1/?report=objectonly HRP C isozymes] the most common ones. <ref name="review">Veitch, N.C. Horseadich peroxidase: a modern view of a classic enzyme 65:249-259 (2004). [https://doi.org/10.1016/j.phytochem.2003.10.022 DOI: 10.1016/j.phytochem.2003.10.022]</ref> |
However, most of the HRP research has focused on one isozyme: <scene name='84/845930/Hrp_c1a/2'>HRP C1A</scene> ([http://www.rcsb.org/structure/1H58 1H58]) <ref name="ref2">Krainer, F.W; GLIEDER, A. An updated view on horseradish peroxidases: recombinant production and biotechnological applications v. 99, pages 1611–1625 (2015). [https://doi.org/10.1007/s00253-014-6346-7 DOI: 10.1007/s00253-014-6346-7]</ref>. | However, most of the HRP research has focused on one isozyme: <scene name='84/845930/Hrp_c1a/2'>HRP C1A</scene> ([http://www.rcsb.org/structure/1H58 1H58]) <ref name="ref2">Krainer, F.W; GLIEDER, A. An updated view on horseradish peroxidases: recombinant production and biotechnological applications v. 99, pages 1611–1625 (2015). [https://doi.org/10.1007/s00253-014-6346-7 DOI: 10.1007/s00253-014-6346-7]</ref>. | ||
| Line 7: | Line 7: | ||
HRP C1A is composed by 308 residues and the residue at <scene name='84/845930/Residue37/1'>position 37</scene> is Ile according to the GenBank entry [http://www.ncbi.nlm.nih.gov/nuccore/M37156.1 M37156.1] but Tyr according to the GenBank entry [http://www.ncbi.nlm.nih.gov/nuccore/HE963800.1 HE963800.1] <ref name="ref2"/>. | HRP C1A is composed by 308 residues and the residue at <scene name='84/845930/Residue37/1'>position 37</scene> is Ile according to the GenBank entry [http://www.ncbi.nlm.nih.gov/nuccore/M37156.1 M37156.1] but Tyr according to the GenBank entry [http://www.ncbi.nlm.nih.gov/nuccore/HE963800.1 HE963800.1] <ref name="ref2"/>. | ||
| - | The molecule has a predominantly α-helical <scene name='84/845930/1h58_secondary_structure/1'>secondary structure</scene>, with the exception of one short β-sheet region, and it is separated into a distal and a proximal region, each one | + | The molecule has a predominantly α-helical <scene name='84/845930/1h58_secondary_structure/1'>secondary structure</scene>, with the exception of one short β-sheet region, and it is separated into a distal and a proximal region, each one has a <scene name='84/845930/Calcio/2'>calcium atom</scene> that is essential in maintaining structural integrity <ref name="review"/>. |
In the center of HRP C1A there is a <scene name='84/845930/Heme/1'>heme group</scene>, which is linked to the molecule by a coordinate bond of the heme iron with a conserved <scene name='84/845930/His170_v2/2'>His170</scene> residue <ref name="review"/>. | In the center of HRP C1A there is a <scene name='84/845930/Heme/1'>heme group</scene>, which is linked to the molecule by a coordinate bond of the heme iron with a conserved <scene name='84/845930/His170_v2/2'>His170</scene> residue <ref name="review"/>. | ||
| - | There are sites for N glycosylation in the loop regions, at <scene name='84/845930/Asnnglicosylation/2'>Asn13, Asn57, Asn58, Asn186, Asn198, Asn214, Asn255 and Asn268</scene> residues<ref name="review"/>. All these glycosylated Asn residues are located on the surface of | + | There are sites for N glycosylation in the loop regions, at <scene name='84/845930/Asnnglicosylation/2'>Asn13, Asn57, Asn58, Asn186, Asn198, Asn214, Asn255 and Asn268</scene> residues<ref name="review"/>. All these glycosylated Asn residues are located on the surface of this enzyme<ref name="ref2"/>. Its carbohydrate content varies but the usual values are between 18% to 22% in mass<ref name="review"/>. |
| - | + | There are also 4 <scene name='84/845930/Saline_bridges_h158/1'>disulphide bridges</scene>: Cys11-Cys91, Cys44-Cys49, Cys97-Cys301 and Cys177-Cys209<ref name="review"/>. | |
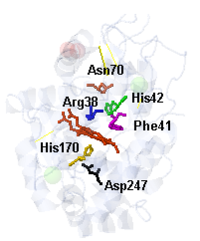
| + | Other residues play essencial roles in the the molecule, as the <scene name='84/845930/Arg38/2'>Arg38</scene> and <scene name='84/845930/Hist42/1'>His42</scene>, which are related to the formation and stabilization of the Compound I in the Peroxidase Cycle <ref name="ref2"/>. | ||
| - | + | His42, Arg38 and <scene name='84/845930/Asn70_1h58_v2/1'>Asn70</scene> are depicted as active agents in the enzyme catalysis. They act as proton transfer agents that enables cleavage of O-O bond of peroxide <ref name="ref2"/>. | |
| - | + | <scene name='84/845930/Phe41_1h58/1'>Phe41</scene> act as a pi-stacking site that binds to aromatic molecules and facilitates its reaction with the iron-site <ref name="ref2"/>. | |
| + | The image below shows the active site in details <ref name="review"/>: | ||
| - | + | [[Image:HRP_C_ACTIVE_SITE.png|200px]] | |
| - | + | == Peroxidase Cycle == | |
| - | <scene name='84/845930/ | + | Peroxidases catalyse the oxidation of a wide variety of substrates, such as phenols, aromatic amines, thioanisoles and iodide, by H2O2. The reaction is a three-step cyclic process, in which the enzyme in its ground state is first oxidised by H2O2 to <scene name='84/845930/Compoundi/1'>Compound I</scene> ([http://www.rcsb.org/structure/1HCH 1HCH]) and then reduced back to the native form in two sequential steps involving a 1 electron reduction and the formation of an enzyme intermediate <scene name='84/845930/Compoundii_v2/1'>Compound II</scene> ([http://www.rcsb.org/structure/1H55 1H55])<ref name="review"/>. Compound I is capable of oxidising a wide range of reducing substrate |
| + | molecules<ref name="ref3">Azevedo AM, Martins VC, Prazeres DM, Vojinović V, Cabral JM, Fonseca LP. Horseradish peroxidase: a valuable tool in biotechnology. Biotechnol Annu Rev. 2003;9:199‐247 [https://doi.org/10.1016/S1387-2656(03)09003-3 DOI: 10.1016/s1387-2656(03)09003-3]</ref>. | ||
| - | < | + | According to the most accepted mechanism to formation of Compound I, the reaction starts when H2O2 binds to the heme iron leading to the formation of a ligand bond between the iron and one of the peroxide oxygens (called Oα) and in the subsequent abstraction of a proton from this oxygen by His42<ref name="ref3"/>. |
| - | < | + | The next step consists in the heterolytic cleavage of the O–O bond and a water molecule is then produced, while one of the oxygens (Oα) remains bonded to the heme iron. In the mechanism described above, the His42 plays the main role as both proton acceptor from Oα and proton donor to the other oxygen (Oβ) and the Arg38 facilitates the cleavage of the O–O bond by promoting charge stabilization<ref name="ref3"/>. |
| - | <scene name='84/845930/ | + | The mechanism of Compounds I and II reduction has not been as intensively studied, but it is generally accepted that when the substrate molecule binds to Compound I, an electron is transferred to the porphyrin ring, It is also known that this reduction is accompanied by the uptake of a proton that has been sugested to bound to the imidazole side chain of His42<ref name="ref3"/>. |
| + | |||
| + | The reduction of Compound II occurs by a similar mechanism but this time the final destination of both proton and electron is the ferryl oxygen. As the ferryl heme iron (Fe IV) is reduced to the ferric state (Fe III), the ferryl oxygen accepts two protons (one from the substrate molecule and the other from the His42 to form a water molecule that is released from the heme iron<ref name="ref3"/>). | ||
| + | |||
| + | [[Image:Horseadish_peroxidase_cycle_v3.jpg|300px]] | ||
| + | |||
| + | == Oxidase Cycle == | ||
| + | |||
| + | Besides the Peroxidase Cycle, HRP C is also known to have an Oxidase Cycle. Its function is not well understood but is related to oxidation/degradation of Indole-3-acetc acid or auxine (IAA), a plant hormone, at acidic pH<ref name="review"/>. This cycle is responsible for the production of the <scene name='84/845930/Compoundiii/1'>Compound III</scene> ([http://www.rcsb.org/structure/1H57 1H57]), which has a hydroperoxyl bounded to the iron<ref name="ref3"/> | ||
| + | |||
| + | [[Image:Horseadish_oxidase_cycle_v2.jpg|300px]] | ||
| + | |||
| + | ==Biological Function == | ||
| + | |||
| + | It has been reported that peroxidases play many different roles in plant life cycle, affecting many processes, such as lignification, pathogen defense, wound healing and Auxin catabolism <ref name="ref4"/>. | ||
| + | |||
| + | [http://en.wikipedia.org/wiki/Lignin Lignin] is a heteropolymer of phenolic monomers. It is present in cell walls and contribute to mechanical strenght and rigidity and chemical and biological resistance. It is formed mainly by four different phenolic monomers, which are oxidized to a phenoxy radical by a peroxidase and further polymerized<ref name="ref4"/>. | ||
| + | |||
| + | Peroxidases also participate in the suberization process. Suberized tissues are highly crosslinked like lignin but are hydrophobic due to being formed by hydroxy fatty acids. It protects the cell from water loss and pathogen invasion by forming a physical barrier to pathogen invasion. These tissue is commonly found in underground organs like roots, where HRP C is found. Peroxidase participate in the polymerization of the monomers by a similar process than lignification <ref name="ref4"/>. | ||
| + | |||
| + | HRP C is involved in the oxidative decarboxylation of [http://en.wikipedia.org/wiki/Auxin auxin] (indole-3-acetic acid or IAA), which is a hormone involved in plant growth regulation. It regulates growth by means of apical dominance, root growth and adventitious growth formation and wound healing. HRP C degrades IAA by different mechanisms, peroxide oxidation by peroxidase cycle and through a oxidase cycle that is independent of peroxide <ref name="ref4"/>. | ||
| + | |||
| + | HRP C level is so a balance between growth and resistance, high levels enables high lignification and subarization protecting the plant but in contrast higher degradation of auxin hinder its growth <ref name="ref4"/>. | ||
| + | |||
| + | |||
| + | ==Biotechnological Uses == | ||
| + | |||
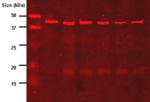
| + | HRP conjugates have been extensively used in immunoassays, such as [http://en.wikipedia.org/wiki/ELISA ELISA] (Enzyme-linked Immunosorbent Assays), [http://en.wikipedia.org/wiki/Western_blot Western-blotting] and [http://en.wikipedia.org/wiki/Immunohistochemistry Immunohistochemistry (IHC)] techniques <ref name="ref3"/>. | ||
| + | |||
| + | [[Image:HRP_ELISA.jpg|150px|right|thumb|ELISA — Enzyme Linked Immuno Sorbent Assay]] | ||
| + | [[Image:HRP_Western_Blotting2.png|150px|right|thumb|Western-blot]] | ||
| + | [[Image:ihc_hrp.jpg|150px|right|thumb|Immunohistochemistry]] | ||
| + | |||
| + | HRP is ideal to the preparation of markers and tracers for immunoassays due to its capability of generate chromogenic products from commercially available substrates with high efficiency and to its stability enabling long term storage <ref name="ref3"/>. | ||
| + | |||
| + | ELISA and Western-blotting use HRP conjugated to antibodies/antigens that bind to the desired species, this way the analyte can be visualised with a indirect HRP chromogenic essay with peroxide and a chromogenic substrate which produces an insoluble colored product <ref name="ref3"/>. | ||
| + | |||
| + | Searching the [http://www.thermofisher.com/br/en/home/life-science/protein-biology/protein-assays-analysis/elisa/elisa-reagents-buffers/enzyme-substrates-elisa.html thermofisher catalog] is possible to find ELISA enzyme-substrate pairs, which most of them currently being HRP-based. | ||
| + | |||
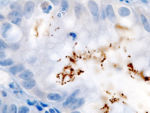
| + | The application in histo- and cytochemistry is also well established, either alone as a protein tracer or conjugated with antibodies for immuno labelling. HRP is one of the most frequently used label for a large variety of molecules used in IHC techniques. In the presence of H2O2, HRP produces detectable products visible by light and electron microscopy, permitting precise cell/tissue localisation <ref name="ref3"/>. | ||
| + | Organic Synthesis. | ||
| + | |||
| + | Enzymes are widely recognised as one of the most promissor catalysts for fine chemical synthesis. They have the ability to produce chemo, stereo and regiospecific chemicals, under mild and environmentally friendly conditions. HRP has proven to be a catalyst to a set of reactions using a green oxidant (peroxide) and to be a chiral derivatizing agent <ref name="ref3"/>. | ||
| + | |||
| + | HRP have been used as a polymerization catalyst for phenols and vinyl monomers, such as acrylamides and methacrylates and for site-specific hydroxylation, such as for L-DOPA (a parkinson’s disease drug), which have been produced using HRP hydroxylation from L-tyrosine (it is not the currently industrial method for its production) and for adrenaline from L-phenylephrine <ref name="ref3"/>. | ||
| + | |||
| + | Also, the product of the reaction between HRP C and auxin generates a citotoxic product that has potencial to be used for cancer treatment<ref name="review"/>. | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Contents |
HORSERADISH PEROXIDASE C1A
|
Horseadish (Armoracia rusticana) is a plant that belongs to the Brassicaceae family. The roots of this plant are rich in class III peroxidases[1], being the HRP C HRP C isozymes the most common ones. [2] However, most of the HRP research has focused on one isozyme: (1H58) [3].
Structural highlights
HRP C1A is composed by 308 residues and the residue at is Ile according to the GenBank entry M37156.1 but Tyr according to the GenBank entry HE963800.1 [3].
The molecule has a predominantly α-helical , with the exception of one short β-sheet region, and it is separated into a distal and a proximal region, each one has a that is essential in maintaining structural integrity [2].
In the center of HRP C1A there is a , which is linked to the molecule by a coordinate bond of the heme iron with a conserved residue [2].
There are sites for N glycosylation in the loop regions, at residues[2]. All these glycosylated Asn residues are located on the surface of this enzyme[3]. Its carbohydrate content varies but the usual values are between 18% to 22% in mass[2].
There are also 4 : Cys11-Cys91, Cys44-Cys49, Cys97-Cys301 and Cys177-Cys209[2].
Other residues play essencial roles in the the molecule, as the and , which are related to the formation and stabilization of the Compound I in the Peroxidase Cycle [3].
His42, Arg38 and are depicted as active agents in the enzyme catalysis. They act as proton transfer agents that enables cleavage of O-O bond of peroxide [3].
act as a pi-stacking site that binds to aromatic molecules and facilitates its reaction with the iron-site [3].
The image below shows the active site in details [2]:
Peroxidase Cycle
Peroxidases catalyse the oxidation of a wide variety of substrates, such as phenols, aromatic amines, thioanisoles and iodide, by H2O2. The reaction is a three-step cyclic process, in which the enzyme in its ground state is first oxidised by H2O2 to (1HCH) and then reduced back to the native form in two sequential steps involving a 1 electron reduction and the formation of an enzyme intermediate (1H55)[2]. Compound I is capable of oxidising a wide range of reducing substrate molecules[4].
According to the most accepted mechanism to formation of Compound I, the reaction starts when H2O2 binds to the heme iron leading to the formation of a ligand bond between the iron and one of the peroxide oxygens (called Oα) and in the subsequent abstraction of a proton from this oxygen by His42[4].
The next step consists in the heterolytic cleavage of the O–O bond and a water molecule is then produced, while one of the oxygens (Oα) remains bonded to the heme iron. In the mechanism described above, the His42 plays the main role as both proton acceptor from Oα and proton donor to the other oxygen (Oβ) and the Arg38 facilitates the cleavage of the O–O bond by promoting charge stabilization[4].
The mechanism of Compounds I and II reduction has not been as intensively studied, but it is generally accepted that when the substrate molecule binds to Compound I, an electron is transferred to the porphyrin ring, It is also known that this reduction is accompanied by the uptake of a proton that has been sugested to bound to the imidazole side chain of His42[4].
The reduction of Compound II occurs by a similar mechanism but this time the final destination of both proton and electron is the ferryl oxygen. As the ferryl heme iron (Fe IV) is reduced to the ferric state (Fe III), the ferryl oxygen accepts two protons (one from the substrate molecule and the other from the His42 to form a water molecule that is released from the heme iron[4]).
Oxidase Cycle
Besides the Peroxidase Cycle, HRP C is also known to have an Oxidase Cycle. Its function is not well understood but is related to oxidation/degradation of Indole-3-acetc acid or auxine (IAA), a plant hormone, at acidic pH[2]. This cycle is responsible for the production of the (1H57), which has a hydroperoxyl bounded to the iron[4]
Biological Function
It has been reported that peroxidases play many different roles in plant life cycle, affecting many processes, such as lignification, pathogen defense, wound healing and Auxin catabolism [1].
Lignin is a heteropolymer of phenolic monomers. It is present in cell walls and contribute to mechanical strenght and rigidity and chemical and biological resistance. It is formed mainly by four different phenolic monomers, which are oxidized to a phenoxy radical by a peroxidase and further polymerized[1].
Peroxidases also participate in the suberization process. Suberized tissues are highly crosslinked like lignin but are hydrophobic due to being formed by hydroxy fatty acids. It protects the cell from water loss and pathogen invasion by forming a physical barrier to pathogen invasion. These tissue is commonly found in underground organs like roots, where HRP C is found. Peroxidase participate in the polymerization of the monomers by a similar process than lignification [1].
HRP C is involved in the oxidative decarboxylation of auxin (indole-3-acetic acid or IAA), which is a hormone involved in plant growth regulation. It regulates growth by means of apical dominance, root growth and adventitious growth formation and wound healing. HRP C degrades IAA by different mechanisms, peroxide oxidation by peroxidase cycle and through a oxidase cycle that is independent of peroxide [1].
HRP C level is so a balance between growth and resistance, high levels enables high lignification and subarization protecting the plant but in contrast higher degradation of auxin hinder its growth [1].
Biotechnological Uses
HRP conjugates have been extensively used in immunoassays, such as ELISA (Enzyme-linked Immunosorbent Assays), Western-blotting and Immunohistochemistry (IHC) techniques [4].
HRP is ideal to the preparation of markers and tracers for immunoassays due to its capability of generate chromogenic products from commercially available substrates with high efficiency and to its stability enabling long term storage [4].
ELISA and Western-blotting use HRP conjugated to antibodies/antigens that bind to the desired species, this way the analyte can be visualised with a indirect HRP chromogenic essay with peroxide and a chromogenic substrate which produces an insoluble colored product [4].
Searching the thermofisher catalog is possible to find ELISA enzyme-substrate pairs, which most of them currently being HRP-based.
The application in histo- and cytochemistry is also well established, either alone as a protein tracer or conjugated with antibodies for immuno labelling. HRP is one of the most frequently used label for a large variety of molecules used in IHC techniques. In the presence of H2O2, HRP produces detectable products visible by light and electron microscopy, permitting precise cell/tissue localisation [4]. Organic Synthesis.
Enzymes are widely recognised as one of the most promissor catalysts for fine chemical synthesis. They have the ability to produce chemo, stereo and regiospecific chemicals, under mild and environmentally friendly conditions. HRP has proven to be a catalyst to a set of reactions using a green oxidant (peroxide) and to be a chiral derivatizing agent [4].
HRP have been used as a polymerization catalyst for phenols and vinyl monomers, such as acrylamides and methacrylates and for site-specific hydroxylation, such as for L-DOPA (a parkinson’s disease drug), which have been produced using HRP hydroxylation from L-tyrosine (it is not the currently industrial method for its production) and for adrenaline from L-phenylephrine [4].
Also, the product of the reaction between HRP C and auxin generates a citotoxic product that has potencial to be used for cancer treatment[2].
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Pandey, Veda & Awasthi, Manika & Singh, Swati & Tiwari, Sameeksha & Dwivedi, Upendra. (2017). A Comprehensive Review on Function and Application of Plant Peroxidases. Biochemistry & Analytical Biochemistry. DOI: 10.4172/2161-1009.1000308
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Veitch, N.C. Horseadich peroxidase: a modern view of a classic enzyme 65:249-259 (2004). DOI: 10.1016/j.phytochem.2003.10.022
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Krainer, F.W; GLIEDER, A. An updated view on horseradish peroxidases: recombinant production and biotechnological applications v. 99, pages 1611–1625 (2015). DOI: 10.1007/s00253-014-6346-7
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 Azevedo AM, Martins VC, Prazeres DM, Vojinović V, Cabral JM, Fonseca LP. Horseradish peroxidase: a valuable tool in biotechnology. Biotechnol Annu Rev. 2003;9:199‐247 DOI: 10.1016/s1387-2656(03)09003-3