User:Felipe de Melo Santana/Sandbox 1
From Proteopedia
| (18 intermediate revisions not shown.) | |||
| Line 6: | Line 6: | ||
== General information == | == General information == | ||
| - | OxyR is a protein that functions as a transcription factor. The OxyR we are analyzing is that of Escherichia Coli. In this perspective, the protein works by regulating the transcription of specific genes, being sensitive to H2O2. This transcription factor belongs to the | + | OxyR is a protein that functions as a transcription factor. The OxyR we are analyzing is that of Escherichia Coli. In this perspective, the protein works by regulating the transcription of specific genes, being sensitive to H2O2.Thus, OxyR acts mainly as a transcriptional activator under oxidizing conditions through direct interaction with the RNA polymerase a subunit.This transcription factor belongs to the Lysr family, with 34 kDa, which forms homotetramers. Under this focus, the members of this family present a domain linked to DNA in their N-terminal region, called helix-loop-helix, a central domain responsible for recognizing H2O2 that is sensitive to the regulation signal and, finally, a domain in C-terminal region responsible for activation and mutimerization. it can be concluded that the OxyR gene encodes a protein that has a dual function, acting as a stress sensor and also as a transcription activator of the OxyR operon. |
| + | |||
== Structure == | == Structure == | ||
| + | |||
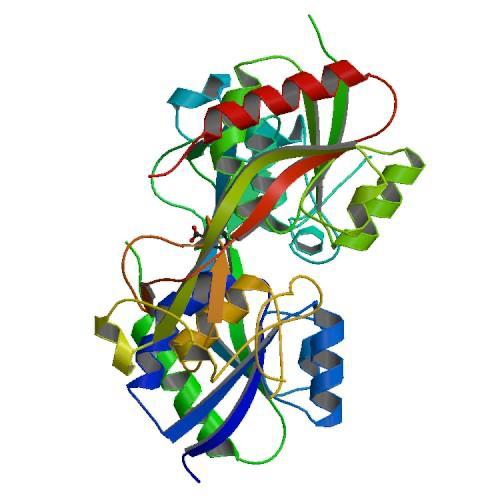
| + | This protein its a transcription factor, at that time its shows two different structures when its reduced or oxidized. The reduced form monomer of the OxyR regulatory domain consists of two α/β domains that are linked by two interdomain strands. Residues <scene name='84/844928/87-162/1'>87–162</scene> in the middle of a folded domain form the majority of the N-terminal a/b domain. The C-terminal a/b Results and Discussion domain II is made of residues <scene name='84/844928/164-260/1'>164–260</scene>. | ||
| + | The remaining C-terminal 30 Structure Determination residues come back to domain I to form an edge of the domain . The two domains exhibit a similar folding pattern consisting of a central b sheet flanked by helices and loops. | ||
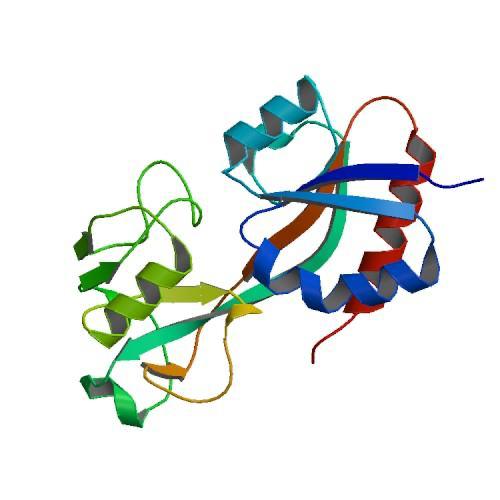
| + | The oxidized form is characterized by the ediction of the domain II, resulting in a significant rearrangement of secondary structures. | ||
| + | |||
| + | [[Image:PHOTO-2020-06-20-17-03-18.jpg]] | ||
| + | |||
| + | Figure 1. CRYSTAL STRUCTURE OF THE REDUCED FORM OF OXYR (1I69) | ||
| + | |||
| + | [[Image:PHOTO-2020-06-20-17-03-26.jpg]] | ||
| + | |||
| + | Figure 2. CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF OXYR (1I64) | ||
== OxyR activation == | == OxyR activation == | ||
| + | |||
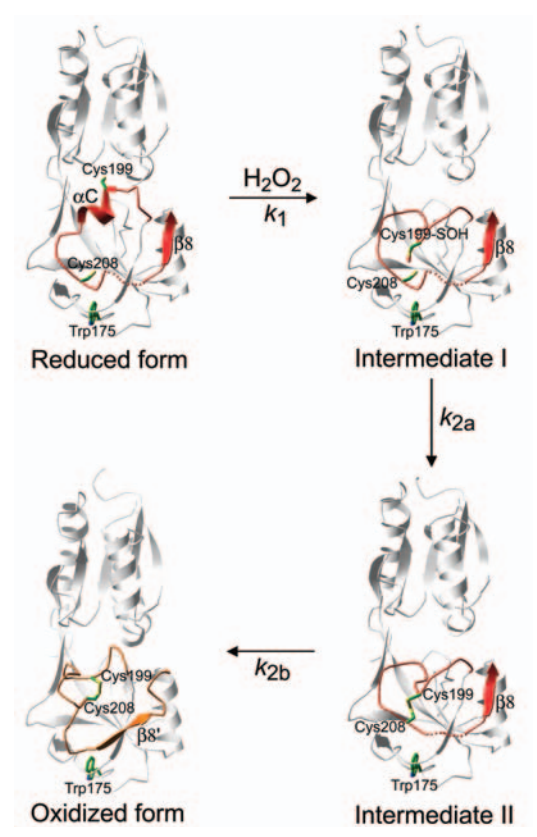
| + | The OxyR transcription factor is activated by the formation of an intramolecular disulfide bond (Zheng et al. 1998). | ||
| + | |||
| + | The OxyR activation, by H2O2, begins with the oxidation of the <scene name='84/844928/Cys_199/1'>Cys199</scene> residue into a sulfenic acid intermediate, which causes destabilization of the Cys199 side chain in the hydrophobic binding pocket, due to its increased size and charge. The destabilization leads to expulsion of the side chain of Cys199 out of the interdomain pocket, resulting in a flexible loop around Cys199 (Lee et al. 2004). | ||
| + | |||
| + | This change in the regulatory domain of OxyR allows the formation of an intramolecular disulfide bond between <scene name='84/844928/Cys_199_-_cys_208/1'>Cys199 and Cys208</scene>, responsible for keeping OxyR in the active form. Thus, OxyR is deactivated by the reduction of the disulfide bond, that leads OxyR back to its reduced form (Zheng et al. 1998). | ||
| + | |||
| + | |||
| + | [[Image:Pasted_image_1.png]] | ||
| + | |||
| + | Figure 3. Intermediates in the structural transitions of OxyR. Lee et al. 2004 | ||
== Protein activity in different forms == | == Protein activity in different forms == | ||
| + | The OxyR transcription factor regulate the expression of genes involved in redox-homeostasis (Stroz et al. 1990). | ||
| - | + | OxyR protein is able to DNA-binding in its two different forms, oxidized and reduced. In your oxidized form, OxyR binds to four adjacent major grooves in DNA, while in the reduced form, OxyR binds to two pairs of adjacent major grooves separated by one helical turn (Toledano et al 1994). These two modes of binding allow OxyR to regulate different promoters, under different cell conditions. | |
| - | OxyR protein is able to DNA-binding in its two different forms, oxidized and reduced. In your oxidized form, OxyR binds to four adjacent major grooves in DNA, while in the reduced form, OxyR binds to two pairs of adjacent major grooves separated by one helical turn ( | + | |
| - | + | ||
| - | + | Both forms of OxyR act as repressors, regulating its own expression by oxyR transcription repression, while only oxidized OxyR is able to activate gene expression, inducing the expression of oxyS (a small regulatory RNA), hydroperoxidase I (katG), alkyl hydroperoxide reductase (ahpCF), glutathione reductase (gorA), glutaredoxin 1 (grxA) and other genes involved in redox-homeostasis, that protect the cell against oxidative stress (Zheng et al. 1998). | |
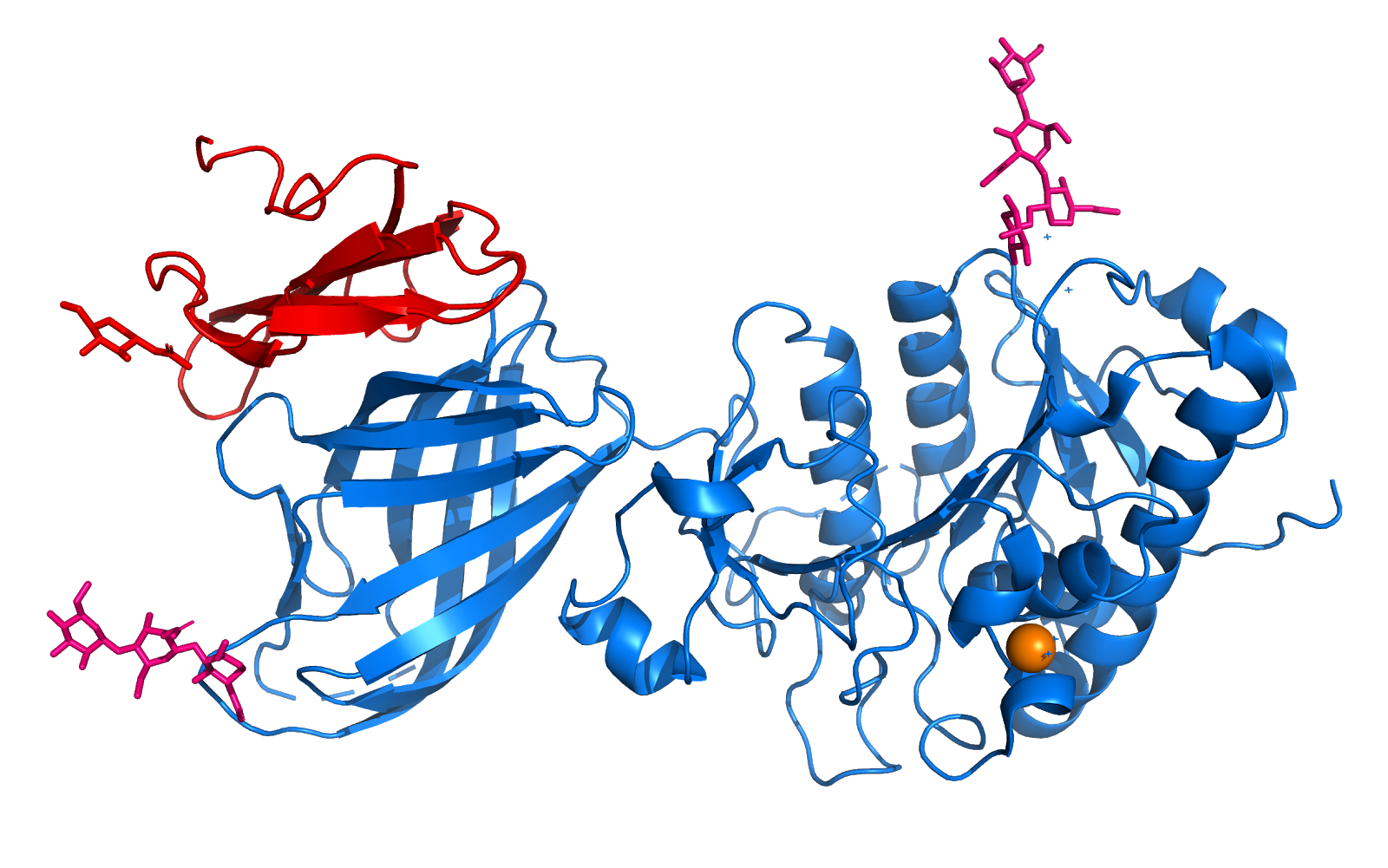
| + | [[Image:Pasted_image_0.png]] | ||
| + | Figure 4. Tetrameric Structure and a Plausible Model of DNA Interactions. Choi et al. 2001 | ||
| + | |||
| + | The tetramers observed in crystals of the reduced (A) and oxidized (B) forms are shown with the DNA positioned on the tetramers with plausible orientations. The tetramers are shown as ribbons (red, green, blue, and yellow for each monomer) and the model of DNA is represented as helical coils (cyan). | ||
| - | == Purification and crystallization == | ||
| Line 33: | Line 59: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | |||
| + | Zheng, M., Aslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721. | ||
| + | |||
| + | Choi H, Kim S, Mukhopadhyay P, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103-113. doi:10.1016/s0092-8674(01)00300-2 | ||
| + | |||
| + | Toledano, M.B., Kullik, I., Trinh, F., Baird, P.T., Schneider, T.D., and Storz, G. (1994). Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909. | ||
| + | |||
| + | Storz, G., Tartaglia, L.A., and Ames, B.N. (1990). Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. | ||
| + | Science 248, 189–194 | ||
| + | |||
| + | Kullik, I., Toledano, M.B., Tartaglia, L.A., and Storz, G. (1995a). Mutational analysis of the redox-sensitive transcriptional regulator OxyR: | ||
| + | regions important for oxidation and transcriptional activation. J. Bacteriol. 177, 1275–1284. | ||
| + | |||
| + | Lee C, Lee SM, Mukhopadhyay P, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11(12):1179-1185. doi:10.1038/nsmb856 | ||
| + | |||
| + | Previato, M. Regulation of genes involved in oxidative stress response in Caulobacter crescentus. 2013. 134 p. Masters thesis (Microbiology) – Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, 2013. | ||
| + | |||
| + | |||
<references/> | <references/> | ||
Current revision
1I69
| |||||||||||
References
Zheng, M., Aslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721.
Choi H, Kim S, Mukhopadhyay P, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103-113. doi:10.1016/s0092-8674(01)00300-2
Toledano, M.B., Kullik, I., Trinh, F., Baird, P.T., Schneider, T.D., and Storz, G. (1994). Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909.
Storz, G., Tartaglia, L.A., and Ames, B.N. (1990). Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248, 189–194
Kullik, I., Toledano, M.B., Tartaglia, L.A., and Storz, G. (1995a). Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177, 1275–1284.
Lee C, Lee SM, Mukhopadhyay P, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11(12):1179-1185. doi:10.1038/nsmb856
Previato, M. Regulation of genes involved in oxidative stress response in Caulobacter crescentus. 2013. 134 p. Masters thesis (Microbiology) – Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, 2013.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644