Journal:Acta Cryst D:S2059798320008475
From Proteopedia
(Difference between revisions)

| (43 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene=' | + | <StructureSection load='' size='450' side='right' scene='85/853720/Cv1/3' caption=''> |
===Getting the best of ID30B for RT data collection in microchips=== | ===Getting the best of ID30B for RT data collection in microchips=== | ||
<big>Jose A. Gavira, Isaac Rodriguez-Ruiz, Sergio Martinez-Rodriguez, Shibom Basu, Sébastien Teychené, Andrew A. McCarthy and Christoph Mueller-Dieckmann</big> <ref>doi 10.1107/S2059798320008475</ref> | <big>Jose A. Gavira, Isaac Rodriguez-Ruiz, Sergio Martinez-Rodriguez, Shibom Basu, Sébastien Teychené, Andrew A. McCarthy and Christoph Mueller-Dieckmann</big> <ref>doi 10.1107/S2059798320008475</ref> | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| + | We have developed a low-cost approach to optimally collect room temperature (RT) data from multiple crystals grown in microchips, using the counter-diffusion technique. We have shown the feasibility of obtaining 3D structural models at the highest attainable resolution, from crystals grown in '''microfluidic systems''', and '''diffracted at RT''' (''i.e.'' '''without harvesting individual crystals'''). Since sample handling and manipulation for cryo-protection are critical factors for X-ray structural determination, RT data collection of the three model proteins, glucose isomerase, lysozyme and thaumatin allow us to determine their corresponding models at atomic resolution, near 1.0 Å. | ||
| + | |||
| + | Furthermore, RT diffraction data can reveal motions crucial for catalysis, ligand binding, and allosteric regulation, not always accessible under standard cryogenic data collection. The '''chips are fabricated by a combination''' of either '''OSTEMER-Kapton''' or '''OSTEMER-Mylar''' materials, both produce a sufficiently low scattering background to permit atomic resolution diffraction data collection at room temperature. The proposed system can be easily incorporated into a fully automatized workflow at any synchrotron beamline facilitating the collection of users' data with no-intervention required of the end-user. | ||
| + | |||
| + | *<scene name='85/853720/Cv1/2'>Glucose isomerase</scene>, Kapton glucose isomerase ([[6ybo]]) is in yellow green; Mylar glucose isomerase ([[6ybr]]) is in cyan. | ||
| + | |||
| + | <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Stop Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | *<scene name='85/853720/Cv1/5'>Lysozyme</scene>, Kapton lysozyme ([[6ybf]]) is in magenta; Mylar lysozyme ([[6ybi]]) is in yellow. | ||
| + | |||
| + | <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Stop Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | *<scene name='85/853720/Cv1/6'>Thaumatin</scene>, Kapton thaumatin ([[6yc5]]) is in salmon; Mylar thaumatin ([[6ybx]]) is in lavender. | ||
| + | |||
| + | <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Stop Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
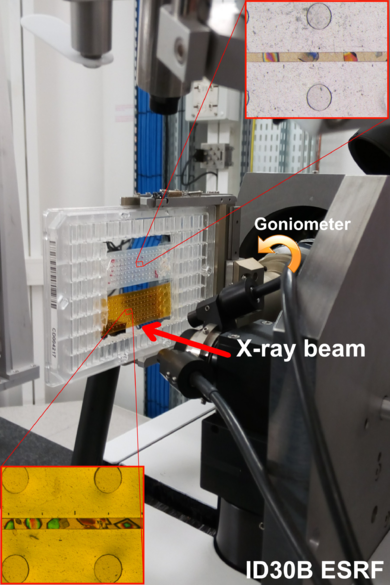
| + | [[Image:Fig1aa.png|thumb|390px|left|Setup mounted for diffraction experiments at the ID30B beamline of the ESRF with two microchips (Kapton and Mylar) hold in a standard crystallization microplate mount in the plate-gripper goniometer head. Insert are images of thaumatin crystals.]] | ||
| + | |||
| + | '''PDB references:''' RT structure of HEW Lysozyme obtained at 1.13 A resolution from crystal grown in a Kapton microchip [[6ybf]]; RT structure of HEW Lysozyme obtained at 1.12 A resolution from crystal grown in a Mylar microchip [[6ybi]]; RT structure of Glucose Isomerase obtained at 1.06 A resolution from crystal grown in a Kapton microchip [[6ybo]]; RT structure of Glucose Isomerase obtained at 1.20 A resolution from crystal grown in a Mylar microchip [[6ybr]]; RT structure of Thaumatin obtained at 1.14 A resolution from crystal grown in a Mylar microchip [[6ybx]]; RT structure of Thaumatin obtained at 1.35 A resolution from crystal grown in a Kapton microchip [[6yc5]]. | ||
<b>References</b><br> | <b>References</b><br> | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.