Journal:Acta Cryst F:S2053230X20010122
From Proteopedia
(Difference between revisions)

(New page: -) |
|||
| (33 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | - | + | <StructureSection load='' size='450' side='right' scene='85/857139/Cv/1' caption=''> |
| + | ===Crystal structure of the extracellular domain of the receptor-like kinase TMK3 from ''Arabidopsis thaliana''=== | ||
| + | <big>Hong Chen, Yanqiong Kong, Jia Chen, Lan Li, Xiushan Li, Feng Yu, Zhenhua Ming</big> <ref>doi 10.1107/S2053230X20010122</ref> | ||
| + | <hr/> | ||
| + | <b>Molecular Tour</b><br> | ||
| + | In this study, we successfully expressed the extracellular domain of TMK3-ECD (residues 25~482) in an insect-cell secretion expression system. We obtained diffraction-quality crystals and determined the three-dimensional structure for TMK3-ECD. The structure was refined to a resolution of 2.06 Å with R<sub>''work''</sub> of 17.69% and R<sub>''free''</sub> of 20.58% (Table 1). Except for 32 residues from the C-terminus and Gln25 from the N-terminus, all remaining residues were well defined and included in the final refined model. The overall structure of the TMK3-LRR exhibits a “L” shape: | ||
| + | *<scene name='85/857139/Cv/10'>Overall structure of TMK3-LRR domain. 1st orientation</scene> (PDB entry [[7brc]]). | ||
| + | *<scene name='85/857139/Cv/11'>Overall structure of TMK3-LRR domain. 2nd orientation</scene>. The LRRNT, LRRs (numbered as indicated) and the non-LRR region of TMK3-LRR are colored in green, whitesmoke and blue, respectively. The three disulfide bonds (Cys54-Cys61, Cys315-Cys323 and Cys353-Cys361) are depicted in yellow. “N” and “C” represent N- and C-terminus, respectively. | ||
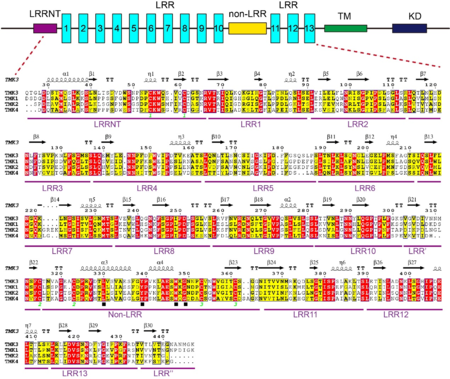
| + | A block of about 40 residues termed non-LRR region intersects the LRR domain into two subdomains – a N-terminal LRR subdomain with 10 LRRs (N-LRRs) and a C-terminal LRR subdomain with 3 LRRs (C-LRRs). C-LRR packs nearly perpendicularly against the spine of N-LRR. Most LRRs from the N-LRR contain the sequence GxL/i/vP (x stands for any amino acid) specific for the plant LRR proteins, with the only exceptions for LRR3, LRR4, LRR7 and LRR10 (see static image below). | ||
| + | |||
| + | [[Image:Tabg.png|thumb|350px|left|Sequence alignment of LRRs in TMK3. The boundary of each LRR is shown on its right. The conserved residues are shown with yellow background. The solid black star indicates the amino acids specific to plant LRR proteins.]] | ||
| + | {{Clear}} | ||
| + | The non-LRR has a Cys cluster with the pattern of ''Cx''<sub>''6-7''</sub>''Cx''<sub>''29-30''</sub>''Cx''<sub>''6-11''</sub>''C'' (Cys315-Cys323 and Cys353-Cys361) and a conserved motif of Lx8Yx7-8WxG (see static image below) similar to Yx8KG found in many LRR- receptor-like kinases (Fritz-Laylin ''et al.,'' 2005<ref name="Fritz">PMID:15955925</ref>). | ||
| + | |||
| + | [[Image:Qw.png|thumb|450px|left|Sequence alignments of the ECDs of TMK3 and its homologs from Arabidopsis. Top: schematic representation of TMK3 protein. TM: transmembrane domain; KD: kinase domain; LRR: leucine-rich repeat. Bottom: Sequence alignment of the ECDs of TMK3 and its homologs from Arabidopsis. Conserved and similar residues are highlighted with red and yellow ground respectively. The solid black square indicates a conserved motif (Lx8Yx7-8WxG). Two cysteines that form a disulfide bond are labeled with the same number (in green) at bottom.]] | ||
| + | {{Clear}} | ||
| + | Most of LRR structures have caps, which shield the hydrophobic core of the first LRR unit at the N-terminus and the last unit at the C-terminus. In extracellular proteins or extracellular regions, the N-terminal and C-terminal caps frequently consist of Cys clusters including two or four Cys residues. The Cys clusters on the N- terminal and C-terminal sides of the LRR arcs are called LRRNT and LRRCT, respectively. Almost all known typical LRR structures include an LRRNT, some of which also have an LRRCT. TMK3-LRR belongs to the LRR subgroups that lacks LRRCT and possesses only an LRRNT (with Cx6C) characterized by a disulfide bond between Cys54 and Cys61. We have also observed <scene name='85/857139/Cv/12'>five asparagine residues (N165, N170, N223, N286 and N448) modified by N-glycosylation</scene>. The N-linked sugars (N-acetylglucosamines) are depicted in magenta sticks. | ||
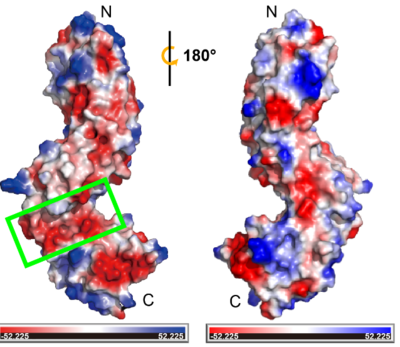
| + | The physiological function of TMK3-ECD is not clearly understood due to the lack of information on the correlation between function and structure. The non-LRR might contribute to ligand interactions (Matsushima ''et al.,'' 2009<ref name="Matsushima">PMID:19580846</ref>; Matsushima & Miyashita, 2012<ref name="Matsushima1">PMID:24970139</ref>). Based on circular dichroism data, Afzal and Lightfoot (Afzal and Lightfoot, 2007<ref name="Afzal">PMID:17287130</ref>) suggested that non-LRR may provide binding diversity to the domains and participate in ligand/protein interactions, dimerization or both. The TMK3-ECD structure revealed a significant electronegative groove located at the non-LRR region between the two LRR domains (see static image below). | ||
| + | |||
| + | [[Image:Elk.png|thumb|400px|left|Two rotated views of the electrostatic surface of TMK3. The green rectangle depicts the non-LRR region.]] | ||
| + | {{Clear}} | ||
| + | This suggests that the non-LRR region is likely a ligand binding pocket that can accommodate positively charged ligands, such as peptides with basic side chains or other electropositive signaling molecules. In addition, the disulfide bonds that stabilize and orient the fold of non-LRR region in TMK3-LRR could be essential for functionality. The disulfide bond between Cys353 to Cys361 is conserved with classical N-cap structure, suggesting its function as an N-cap to stabilize the structure of C-LRR. The other disulfide bond (Cys315-Cys323) is responsible for connecting two LRR domains. Breaking this pairs of disulfide bond may cause the two LRR domains to be converted into one, altering the ligand binding interaction. Besides, it may also play a critical role in a changing redox environment. The number of N-LRR is greater than the number of C-LRR, which brings close proximity of the non-LRR to interact with ligand and the transmembrane region. It also might facilitate signaling in the cytoplasm through interactions with ligand. We also speculate it's possible to have a role for the N- LRR and C-LRR domains that provides more than one scaffold for protein interaction or ligand recognition. | ||
| + | |||
| + | <scene name='85/857139/Cv/13'>TMK3-LRR structure alignment of the TMK1-LRR are shown in blue (TMK1) and red (TMK3)</scene>. | ||
| + | |||
| + | <scene name='85/857139/Cv/14'>Alignment with TMK3-LRR, BRI1-LRR and PSYR-LRR</scene>. The structure of the TMK3-LRR, BRI1-LRR and PSYR-LRR are depicted in red, blue and yellow. | ||
| + | |||
| + | '''PDB reference:''' extracellular domain of TMK3, [[7brc]]. | ||
| + | |||
| + | <b>References</b><br> | ||
| + | <references/> | ||
| + | </StructureSection> | ||
| + | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.