Histone Lysine Methyltransferase SET7/9
From Proteopedia
(Difference between revisions)
(Category:Featured in BAMBED. Date must be of the page, not the paper) |
|||
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | {{BAMBED | ||
| + | |DATE=March 10, 2022 | ||
| + | |OLDID=3412167 | ||
| + | |BAMBEDDOI=10.1002/bmb.21759 | ||
| + | }} | ||
=SET7/9, A Histone Lysine Methyltransferase and epigenetic activator of transcription= | =SET7/9, A Histone Lysine Methyltransferase and epigenetic activator of transcription= | ||
| Line 7: | Line 12: | ||
===Histone Methylation=== | ===Histone Methylation=== | ||
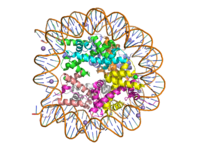
[[Image:human_nucleosome_ray_trace.png|200 px|right|thumb|Figure 1: Human nucleosome particle, pbd code: 5y0c]] | [[Image:human_nucleosome_ray_trace.png|200 px|right|thumb|Figure 1: Human nucleosome particle, pbd code: 5y0c]] | ||
| - | [https:// | + | [https://proteopedia.org/wiki/index.php/Nucleosome Histone proteins] aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The nucleosome (Figure 1) itself is an octamer complex of the core histone proteins [https://en.wikipedia.org/wiki/Histone_H2A H2a], [https://en.wikipedia.org/wiki/Histone_H2B H2b], [https://en.wikipedia.org/wiki/Histone_H3 H3], and [https://en.wikipedia.org/wiki/Histone_H4 H4]. Approximately 145-157 DNA base pairs wind around a histone core and the DNA linking the core particles are bound by [https://en.wikipedia.org/wiki/Histone_H1 Histones H1 and H5].<ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of [https://en.wikipedia.org/wiki/Transcription_factor transcription factors] to the genome, either promoting or inhibiting [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. Some of these modifications include [https://en.wikipedia.org/wiki/Histone_methylation methylation]/[https://en.wikipedia.org/wiki/Demethylation#In_biochemistry demethylation], [https://en.wikipedia.org/wiki/Histone_acetylation_and_deacetylation acetylation/deacetylation], and [https://en.wikipedia.org/wiki/Ubiquitin ubiquitination/deubiquitination].<ref name="Lu">Lu, D. Epigenetic modification enzymes: catalytic mechanisms and inhibitors, ''Acta Pharmaceutica Sinica B.''2013 May. 3(3):141-9 [https://doi.org/10.1016/j.apsb.2013.04.007 DOI:10.1016/j.apsb.2013.04.007]</ref> |
| - | Histone methylation is associated with both gene activation and silencing. The factors influencing the specific outcome are the methylation site, residue type (Lys or Arg) and the extent (mono-, di- or tri-methylation).<ref name="Dong">PMID: 23566087</ref> Sites known for gene activation are Lys-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys-27 on H3 and Lys-20 on H4 are known for transcriptional repression or silencing.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are found on both active and inactive genes. Some tumor related genes such as [https:// | + | Histone methylation is associated with both gene activation and silencing. The factors influencing the specific outcome are the methylation site, residue type (Lys or Arg) and the extent (mono-, di- or tri-methylation).<ref name="Dong">PMID: 23566087</ref> Sites known for gene activation are Lys-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys-27 on H3 and Lys-20 on H4 are known for transcriptional repression or silencing.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are found on both active and inactive genes. Some tumor related genes such as [https://proteopedia.org/wiki/index.php/P53 p53] are site specifically methylated to promote their biological function <ref name = "Rizzo" />, whereas hypomethylation of [https://en.wikipedia.org/wiki/CpG_site#Methylation,_silencing,_cancer,_and_aging CpG island sequences] is linked to tumor genesis.<ref name="Lu" /> |
| - | Sidechain methylation of basic amino acids in histone proteins is accomplished by the Histone Methyltransferase class of enzymes. In all but one instance, the many enzymes that are lysine substrate specific contain the structural protein domain known as the [https://en.wikipedia.org/wiki/SET_domain SET domain]. Generally, these enzymes use S- | + | Sidechain methylation of basic amino acids in histone proteins is accomplished by the Histone Methyltransferase class of enzymes. In all but one instance, the many enzymes that are lysine substrate specific contain the structural protein domain known as the [https://en.wikipedia.org/wiki/SET_domain SET domain]. Generally, these enzymes use S-adenosyl methionine to methylate lysine in the H3, H4 and H2a core proteins. One enzyme in this family is the <scene name='83/833386/Kmt_full/8'>SET7/9 lysine methyltransferase</scene>, which adds a methyl group to Lys4 of the H3 protein. This histone modification results in the formation of euchromatin and gene transcription.<ref name="Xiao"> PMID:12540855 </ref>, <ref name="Dong" /> |
==Lysine Methyltransferase (KMT) Structure== | ==Lysine Methyltransferase (KMT) Structure== | ||
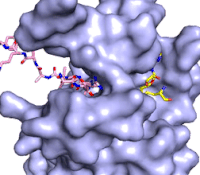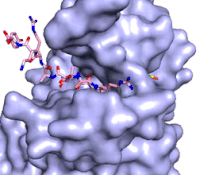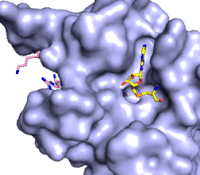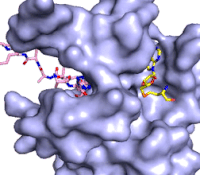
The structure of human histone methyltransferase SET7/9 was determined by x-ray diffraction at 1.75Å resolution. In this structure, a 10-residue peptide representing histone H3 was co-crystallized with SET7/9 and its co-factor product, S-adenosyl homocysteine ([https://en.wikipedia.org/wiki/S-Adenosyl-L-homocysteine SAH]) <ref name="Xiao" />. Here, the histone H3 peptide is methylated at lysine four, representing the product of the reaction. | The structure of human histone methyltransferase SET7/9 was determined by x-ray diffraction at 1.75Å resolution. In this structure, a 10-residue peptide representing histone H3 was co-crystallized with SET7/9 and its co-factor product, S-adenosyl homocysteine ([https://en.wikipedia.org/wiki/S-Adenosyl-L-homocysteine SAH]) <ref name="Xiao" />. Here, the histone H3 peptide is methylated at lysine four, representing the product of the reaction. | ||
| - | + | ||
===Overall Structure=== | ===Overall Structure=== | ||
The SET7/9 enzyme structure sequentially consists of a N-terminal domain (177-193), followed by the characteristic [https://en.wikipedia.org/wiki/SET_domain SET domain] (<scene name='83/833386/Set7_domain/2'>residues 194-343</scene>) which itself ends with a specific C-terminal segment (344-366). The enzyme is best characterized as having [https://en.wikipedia.org/wiki/Protein_fold_class#%CE%B1+%CE%B2_proteins alpha+beta] folding topology as it consists of a mixture of both α-helix and β-sheet, but without any significant repeating pattern <ref name="Xiao" />. The helical composition includes three <scene name='83/833386/Alpha_helices/1'>α-helices</scene>, with two residing in the SET domain and one in the C-terminal segment. The α-helices in the SET domain are two turns in length while the C-terminal helix is by far the largest with four turns. There are also two <scene name='83/833386/3-10_helices/1'>3-10 helices</scene> in the SET domain which are each one turn. There are 21 total <scene name='83/833386/Beta_sheets/2'>β-strands</scene> found in both the N-terminal and the SET domains. The β-strands are primarily anti-parallel and multiple β-strands are connected by Type 1 and Type 2 <scene name='83/833386/Beta_turns/1'>β-turns</scene>. | The SET7/9 enzyme structure sequentially consists of a N-terminal domain (177-193), followed by the characteristic [https://en.wikipedia.org/wiki/SET_domain SET domain] (<scene name='83/833386/Set7_domain/2'>residues 194-343</scene>) which itself ends with a specific C-terminal segment (344-366). The enzyme is best characterized as having [https://en.wikipedia.org/wiki/Protein_fold_class#%CE%B1+%CE%B2_proteins alpha+beta] folding topology as it consists of a mixture of both α-helix and β-sheet, but without any significant repeating pattern <ref name="Xiao" />. The helical composition includes three <scene name='83/833386/Alpha_helices/1'>α-helices</scene>, with two residing in the SET domain and one in the C-terminal segment. The α-helices in the SET domain are two turns in length while the C-terminal helix is by far the largest with four turns. There are also two <scene name='83/833386/3-10_helices/1'>3-10 helices</scene> in the SET domain which are each one turn. There are 21 total <scene name='83/833386/Beta_sheets/2'>β-strands</scene> found in both the N-terminal and the SET domains. The β-strands are primarily anti-parallel and multiple β-strands are connected by Type 1 and Type 2 <scene name='83/833386/Beta_turns/1'>β-turns</scene>. | ||
| Line 27: | Line 32: | ||
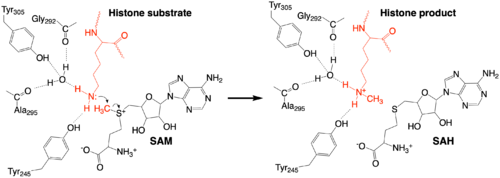
The methylation reaction (Figure 4) is activated by several hydrogen bond interactions between active site residues and the substrate lysine. Specifically, the Tyr305 hydroxyl and the mainchain carbonyl oxygen of both Ala295 and Gly292 coordinate a buried water molecule that in turn coordinates a hydrogen of the lysine substrate. Additionally, the hydroxyl of Tyr245 also hydrogen bonds to the lysine substrate. These interactions enhance the nucleophilic nature of the amine nitrogen so that subsequent attack of the SAM methyl group carbon becomes favorable. Attack is further facilitated as the donor methyl is bound to a positively charged sulfur which will make a good leaving group when demthylated. Once the methyl group is transferred to the amine, the charge on the sulfur is resolved and thus SAM is converted to SAH.<ref name="Xiao" /> | The methylation reaction (Figure 4) is activated by several hydrogen bond interactions between active site residues and the substrate lysine. Specifically, the Tyr305 hydroxyl and the mainchain carbonyl oxygen of both Ala295 and Gly292 coordinate a buried water molecule that in turn coordinates a hydrogen of the lysine substrate. Additionally, the hydroxyl of Tyr245 also hydrogen bonds to the lysine substrate. These interactions enhance the nucleophilic nature of the amine nitrogen so that subsequent attack of the SAM methyl group carbon becomes favorable. Attack is further facilitated as the donor methyl is bound to a positively charged sulfur which will make a good leaving group when demthylated. Once the methyl group is transferred to the amine, the charge on the sulfur is resolved and thus SAM is converted to SAH.<ref name="Xiao" /> | ||
| + | [[Image:KMT_mechanism_final.png|500px|center|thumb|Figure 4: The proposed KMT Mechanism. The lysine substrate and transferred methyl group are in red.]] | ||
===The C-Terminal Domain=== | ===The C-Terminal Domain=== | ||
| - | The C-terminal domain of lysine methyltransferase consists of a β-hairpin and α-helix that serve as a 'cap' for the SET domain. The overall structure of the <scene name='83/833386/C_terminal_domain/1'>C-terminal domain (residues 340-366)</scene> provides various interactions that facilitate binding of substrate to the SET domain (residues 193-344).<ref name="Xiao" /> Hydrophobic interactions between the C-terminal domain and the SET domain are mainly responsible in forming the access channel for the substrate and assist in deprotonation of the lysine. Residues 337-349 create a pro-gly rich <scene name='83/833386/Beta-hairpin/5'>β-hairpin</scene> that stabilizes the orientation of two tyrosine residues, Tyr 335 and Tyr337, that form the lysine access channel. Furthermore, there is substantial hydrophobic packing of the C-terminal helix against the SET domain using <scene name='83/833386/C_terminal_domain/4'>residues Phe299, Tyr353, Leu357 and Phe360 </scene>. These interactions position the indole ring of Trp352 in the C-terminal helix to <scene name='83/833386/C_terminal_domain/6'>stack with the π-cloud of the adenine base of the SAM co-factor</scene> as well as allowing Glu356 to hydrogen bond with N6 of the adenine ring.<ref name="Xiao" /> | + | The C-terminal domain of lysine methyltransferase consists of a β-hairpin and α-helix that serve as a 'cap' for the SET domain. The overall structure of the <scene name='83/833386/C_terminal_domain/1'>C-terminal domain (residues 340-366)</scene> provides various interactions that facilitate binding of substrate to the SET domain (residues 193-344).<ref name="Xiao" /> Hydrophobic interactions between the C-terminal domain and the SET domain are mainly responsible in forming the access channel for the substrate and assist in deprotonation of the lysine. Residues 337-349 create a pro-gly rich <scene name='83/833386/Beta-hairpin/5'>β-hairpin</scene> that stabilizes the orientation of two tyrosine residues, Tyr 335 and Tyr337, that form the lysine access channel. Furthermore, there is substantial hydrophobic packing of the C-terminal helix against the SET domain using <scene name='83/833386/C_terminal_domain/4'>residues Phe299, Tyr353, Leu357 and Phe360 </scene>. These interactions position the indole ring of Trp352 in the C-terminal helix to <scene name='83/833386/C_terminal_domain/6'>stack with the π-cloud of the adenine base of the SAM co-factor</scene> as well as allowing Glu356 to hydrogen bond with N6 of the adenine ring.<ref name="Xiao" /> |
==Inhibitors== | ==Inhibitors== | ||
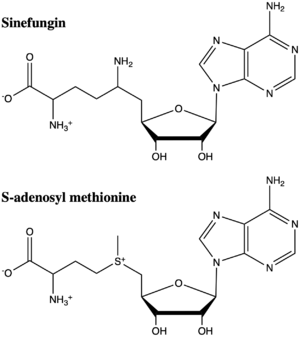
| Line 42: | Line 48: | ||
Another inhibitor of SET7/9 is [https://en.wikipedia.org/wiki/Cyproheptadine cyproheptadine], a clinically-approved anti-allergy drug that was originally developed as histamine (H1) and serotonin (5-HT2A) receptor antagonist.<ref name="Takemoto" /> The structure of <scene name='83/833386/Cyproheptadine/3'>cyproheptadine-SET7/9 complex</scene> bound with the SAM co-factor was determined by X-ray diffraction at 2.0Å resolution (PDB:5ayf). Unlike Sinefungin, which mimics the SAM co-factor, cyproheptadine is a traditional inhibitor that competes with the histone peptide substrate for the peptide-binding site. When cyproheptadine binds to the substrate site, the methylated nitrogen of the [https://en.wikipedia.org/wiki/Piperidine piperdine] ring forms a hydrogen bond with the carbonyl of Thr266 as well as stacking with Tyr335. The binding of cyproheptadine to the catalytic site displaces residue Tyr337, an important residue for the formation of the lysine access channel and unresolved in the inhibited structure. This movement subsequently causes a conformational change of the <scene name='83/833386/1o9s_5ayf_alignment/7'>beta hairpin</scene> rendering it disordered in the structure, and shifts the position of the C-terminal helix.<ref name="Takemoto" /> | Another inhibitor of SET7/9 is [https://en.wikipedia.org/wiki/Cyproheptadine cyproheptadine], a clinically-approved anti-allergy drug that was originally developed as histamine (H1) and serotonin (5-HT2A) receptor antagonist.<ref name="Takemoto" /> The structure of <scene name='83/833386/Cyproheptadine/3'>cyproheptadine-SET7/9 complex</scene> bound with the SAM co-factor was determined by X-ray diffraction at 2.0Å resolution (PDB:5ayf). Unlike Sinefungin, which mimics the SAM co-factor, cyproheptadine is a traditional inhibitor that competes with the histone peptide substrate for the peptide-binding site. When cyproheptadine binds to the substrate site, the methylated nitrogen of the [https://en.wikipedia.org/wiki/Piperidine piperdine] ring forms a hydrogen bond with the carbonyl of Thr266 as well as stacking with Tyr335. The binding of cyproheptadine to the catalytic site displaces residue Tyr337, an important residue for the formation of the lysine access channel and unresolved in the inhibited structure. This movement subsequently causes a conformational change of the <scene name='83/833386/1o9s_5ayf_alignment/7'>beta hairpin</scene> rendering it disordered in the structure, and shifts the position of the C-terminal helix.<ref name="Takemoto" /> | ||
With the revelation of its inhibitory effects on SET7/9, cyproheptadine was used in vitro to treat [https://en.wikipedia.org/wiki/MCF-7 MCF-7] breast cancer cells. SET7/9's non-histone activities include the methylation of the estrogen receptor α ([https://en.wikipedia.org/wiki/Estrogen_receptor_alpha ERα]), a nuclear receptor and a transcription factor responsible for estrogen-responsive gene regulation. The expression and transcriptional activity of ERα is involved in the carcinogenesis of 70% of breast cancers, making it a major target for hormone therapy. Researchers found that treating the MCF-7 cells with cyproheptadine decreased ERα's expression and transcriptional activity which therefore inhibited the estrogen-dependent cell growth. These findings suggest that cyproheptadine could possibly be repurposed to breast cancer therapy in the future.<ref name="Takemoto" /> | With the revelation of its inhibitory effects on SET7/9, cyproheptadine was used in vitro to treat [https://en.wikipedia.org/wiki/MCF-7 MCF-7] breast cancer cells. SET7/9's non-histone activities include the methylation of the estrogen receptor α ([https://en.wikipedia.org/wiki/Estrogen_receptor_alpha ERα]), a nuclear receptor and a transcription factor responsible for estrogen-responsive gene regulation. The expression and transcriptional activity of ERα is involved in the carcinogenesis of 70% of breast cancers, making it a major target for hormone therapy. Researchers found that treating the MCF-7 cells with cyproheptadine decreased ERα's expression and transcriptional activity which therefore inhibited the estrogen-dependent cell growth. These findings suggest that cyproheptadine could possibly be repurposed to breast cancer therapy in the future.<ref name="Takemoto" /> | ||
| - | |||
== References == | == References == | ||
<references/> | <references/> | ||
| - | |||
</StructureSection> | </StructureSection> | ||
| Line 54: | Line 58: | ||
Alexandra Pentala, | Alexandra Pentala, | ||
Madeleine Wilson | Madeleine Wilson | ||
| + | |||
| + | [[Category:Featured in BAMBED]] | ||
Current revision
This page, as it appeared on March 10, 2022, was featured in this article in the journal Biochemistry and Molecular Biology Education.
SET7/9, A Histone Lysine Methyltransferase and epigenetic activator of transcription
| |||||||||||
Student Contributors
Lauren Allman, Lauryn Padgett, Alexandra Pentala, Madeleine Wilson
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Michal Harel, Valentine J Klimkowski, Angel Herraez