We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
EPSP synthase
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
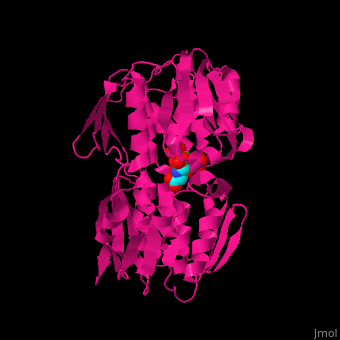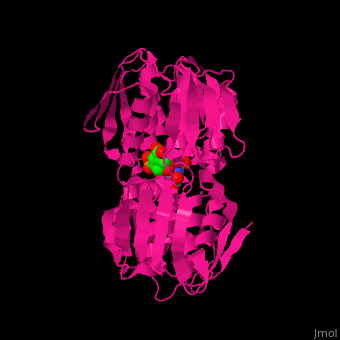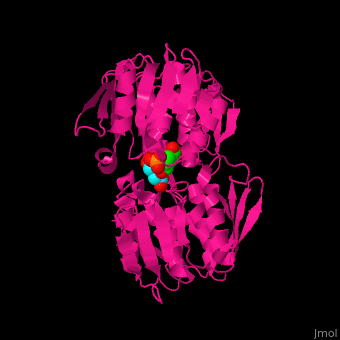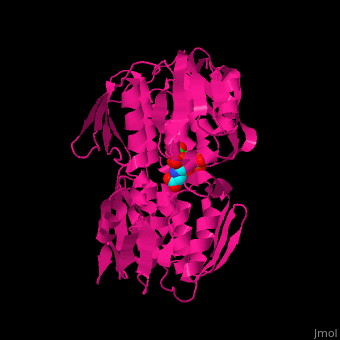
<StructureSection load='' size='350' side='right' caption='Structure of E. coli EPSP synthase complex with shikimate-3-phosphate, the herbicide glyphosate and formic acid (PDB entry [[1g6s]])' scene='57/570585/Cv/1'> | <StructureSection load='' size='350' side='right' caption='Structure of E. coli EPSP synthase complex with shikimate-3-phosphate, the herbicide glyphosate and formic acid (PDB entry [[1g6s]])' scene='57/570585/Cv/1'> | ||
| + | |||
| + | __TOC__ | ||
== Function == | == Function == | ||
| Line 7: | Line 9: | ||
The enzyme has two domains, with the active site found in the interdomain cleft <scene name='57/570585/Two_domains/5'>(open conformation)</scene>. There is a substantial structural change upon substrate binding, resulting in a <scene name='57/570585/Closed_formation/3'>closed</scene> conformation. <scene name='57/570585/Cv/9'>See animation of this process</scene>. '''Glyphosate''' (also known as '''Roundup''') occupies the <scene name='57/570585/Glyphosate_s3p/1'>binding site</scene> of the second substrate, phosphoenol pyruvate <ref>PMID:11171958</ref>. Interestingly CP4 EPSP synthase still binds glyphosate in the absence of PEP, but a conformational change in glyphosate to accommodate a steric clash with <scene name='57/570585/Glyphosate_s3p_distance/2'>Glu 354</scene> shortens the length of glyphosate, from 7.3 angstroms to 6.67 angstroms, and changes the IC50 by a factor of over 4,000, from 2.5 micromolar to 11 millimolar. | The enzyme has two domains, with the active site found in the interdomain cleft <scene name='57/570585/Two_domains/5'>(open conformation)</scene>. There is a substantial structural change upon substrate binding, resulting in a <scene name='57/570585/Closed_formation/3'>closed</scene> conformation. <scene name='57/570585/Cv/9'>See animation of this process</scene>. '''Glyphosate''' (also known as '''Roundup''') occupies the <scene name='57/570585/Glyphosate_s3p/1'>binding site</scene> of the second substrate, phosphoenol pyruvate <ref>PMID:11171958</ref>. Interestingly CP4 EPSP synthase still binds glyphosate in the absence of PEP, but a conformational change in glyphosate to accommodate a steric clash with <scene name='57/570585/Glyphosate_s3p_distance/2'>Glu 354</scene> shortens the length of glyphosate, from 7.3 angstroms to 6.67 angstroms, and changes the IC50 by a factor of over 4,000, from 2.5 micromolar to 11 millimolar. | ||
| - | </StructureSection> | ||
| - | + | ==3D structures of EPSP synthase == | |
| - | + | [[EPSP synthase 3D structures]] | |
| - | + | </StructureSection> | |
| - | *5-enolpyruvylshikimate 3-phosphate (EPSP) synthase | ||
| - | |||
| - | **[[1eps]] – EcEPSP – ''Escherichia coli''<br /> | ||
| - | **[[1p88]], [[1p89]] - EcEPSP N terminal - NMR<br /> | ||
| - | **[[1rf5]] - SpEPSP – ''Streptococcus pneumoniae''<br /> | ||
| - | **[[2bjb]], [[2o15]] – MtEPSP – ''Mycobacterium tuberculosis''<br /> | ||
| - | **[[2gg4]] – AgEPSP – ''Agrobacterium''<br /> | ||
| - | **[[3roi]], [[3tr1]], [[4gfp]] – CbEPSP – ''Coxiella burnetii''<br /> | ||
| - | **[[3ti2]] - VcEPSP N terminal – ''Vibrio cholerae''<br /> | ||
| - | **[[3rmt]] – EPSP – ''Bacillus halodurans''<br /> | ||
| - | |||
| - | *EPSP synthase binary complex | ||
| - | |||
| - | **[[1g6t]] - EcEPSP + S3P <br /> | ||
| - | **[[1mi4]], [[2qfq]], [[2qfs]], [[3fjx]], [[3fk0]] - EcEPSP (mutant) + S3P <br /> | ||
| - | **[[1q36]] - EcEPSP (mutant) + S3P derivative<br /> | ||
| - | **[[1x8r]], [[1x8t]] - EcEPSP + intermediate <br /> | ||
| - | **[[2aa9]] – EcEPSP + shikimate<br /> | ||
| - | **[[2pq9]] - EcEPSP + intermediate <br /> | ||
| - | **[[1rf4]] - SpEPSP + intermediate <br /> | ||
| - | **[[2o0b]], [[2o0d]] - MtEPSP + S3P <br /> | ||
| - | **[[2gg6]] - AgEPSP + S3P <br /> | ||
| - | **[[2pqb]], [[2pqc]], [[2pqd]] - AgEPSP + intermediate <br /> | ||
| - | **[[2o0x]] - MtEPSP + intermediate <br /> | ||
| - | **[[2o0z]] - MtEPSP + EPS <br /> | ||
| - | **[[4egr]] - CbEPSP + phosphoenolpyruvate<br /> | ||
| - | |||
| - | *EPSP synthase ternary complex | ||
| - | |||
| - | **[[1g6s]] - EcEPSP + S3P + glyphosate<br /> | ||
| - | **[[2qft]], [[2qfu]], [[3fjz]], [[3fk1]] - EcEPSP (mutant) + S3P + glyphosate<br /> | ||
| - | **[[2aay]] - EcEPSP + shikimate + glyphosate<br /> | ||
| - | **[[1rf6]] - SpEPSP + S3P + glyphosate<br /> | ||
| - | **[[2gga]] - AgEPSP + S3P + glyphosate<br /> | ||
| - | **[[2ggd]] - AgEPSP (mutant) + S3P + glyphosate<br /> | ||
| - | **[[2o0e]] - MtEPSP + S3P + phosphoenolpyruvate<br /> | ||
| - | **[[3nvs]] - VcEPSP + S3P + glyphosate <br /> | ||
| - | **[[3slh]] - CbEPSP + S3P + glyphosate<br /> | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | |||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Priestman MA, Healy ML, Funke T, Becker A, Schonbrunn E. Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate. FEBS Lett. 2005 Oct 24;579(25):5773-80. PMID:16225867 doi:10.1016/j.febslet.2005.09.066
- ↑ Funke T, Han H, Healy-Fried ML, Fischer M, Schonbrunn E. Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13010-5. Epub 2006 Aug 17. PMID:16916934
- ↑ Schonbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JN, Kabsch W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1376-80. PMID:11171958 doi:http://dx.doi.org/10.1073/pnas.98.4.1376
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Ann Taylor, Joel L. Sussman