We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:R. Jeremy Johnson/Mitochondrial Calcium Uniporter
From Proteopedia
< User:R. Jeremy Johnson(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
== Overview == | == Overview == | ||
| - | The mitochondrial calcium uniporter (MCU) complex is the main source of entry for [https://en.wikipedia.org/wiki/Calcium calcium] ions into the [https://en.wikipedia.org/wiki/Mitochondrial_matrix mitochondrial matrix] from the [https://en.wikipedia.org/wiki/Mitochondrion#Intermembrane_space intermembrane space]. MCU channels exist in most [https://en.wikipedia.org/wiki/Eukaryote eukaryotes], but activity is regulated differently in each [https://en.wikipedia.org/wiki/Clade clade].<ref name="Baradaran">PMID:29995857</ref> MCU was definitively assigned in 2011 using a combination of [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance_spectroscopy NMR spectroscopy], [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy | + | The mitochondrial calcium uniporter (MCU) complex is the main source of entry for [https://en.wikipedia.org/wiki/Calcium calcium] ions into the [https://en.wikipedia.org/wiki/Mitochondrial_matrix mitochondrial matrix] from the [https://en.wikipedia.org/wiki/Mitochondrion#Intermembrane_space intermembrane space]. MCU channels exist in most [https://en.wikipedia.org/wiki/Eukaryote eukaryotes], but activity is regulated differently in each [https://en.wikipedia.org/wiki/Clade clade].<ref name="Baradaran">PMID:29995857</ref> MCU was definitively assigned in 2011 using a combination of [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance_spectroscopy NMR spectroscopy], [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryoelectron microscopy], and [https://en.wikipedia.org/wiki/X-ray_crystallography x-ray crystallography].<ref name="Woods">PMID:31869674</ref> Recent [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryoelectron microscopy] (cryo-EM) analysis provides a structural framework for understanding the mechanism for calcium selectivity by the MCU.<ref name="Giorgi" /> Like other ion channels, the MCU is highly selective and efficient, allowing calcium ions into the mitochondrial matrix at a rate of 5,000,000 ions per second, even though [https://en.wikipedia.org/wiki/Potassium potassium] ions are over 100,000 times more abundant in the intermembrane space.<ref name="Baradaran"/> |
Under resting conditions, the calcium concentration in the mitochondria is about the same as in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm], but when stimulated, mitochondrial calcium concentration increases 10 to 20-fold.<ref name="Giorgi">PMID:30143745</ref> [https://en.wikipedia.org/wiki/Mitochondria_associated_membranes Mitochondria-associated ER membranes] exist between the mitochondria and the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum endoplasmic reticulum] facilitate efficient transport of calcium ions.<ref name="Wang">PMID:28882140</ref> The transfer of electrons through [https://en.wikipedia.org/wiki/Electron_transport_chain#Mitochondrial_redox_carriers respiratory complexes I-IV] produces the energy to pump [https://en.wikipedia.org/wiki/Hydrogen_ion hydrogen ions] into the intermembrane space and establish the proton [https://en.wikipedia.org/wiki/Electrochemical_gradient electrochemical gradient] potential.<ref name="Giorgi"/> This negative electrochemical potential is the driving force that moves positively charged calcium ions into the mitochondrial matrix.<ref name="Giorgi"/> Calcium uptake and efflux must be tightly regulated to controll essential [https://en.wikipedia.org/wiki/Citric_acid_cycle Krebs cycle] enzyme activity, including [http://proteopedia.org/wiki/index.php/Pyruvate_dehydrogenase pyruvate dehydrogenase], [https://en.wikipedia.org/wiki/Oxoglutarate_dehydrogenase_complex α-ketoglutarate dehydrogenase], and [http://proteopedia.org/wiki/index.php/Isocitrate_dehydrogenase isocitrate dehydrogenase], while avoiding calcium overload and [https://en.wikipedia.org/wiki/Apoptosis apoptosis].<ref name="Wang"/> | Under resting conditions, the calcium concentration in the mitochondria is about the same as in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm], but when stimulated, mitochondrial calcium concentration increases 10 to 20-fold.<ref name="Giorgi">PMID:30143745</ref> [https://en.wikipedia.org/wiki/Mitochondria_associated_membranes Mitochondria-associated ER membranes] exist between the mitochondria and the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum endoplasmic reticulum] facilitate efficient transport of calcium ions.<ref name="Wang">PMID:28882140</ref> The transfer of electrons through [https://en.wikipedia.org/wiki/Electron_transport_chain#Mitochondrial_redox_carriers respiratory complexes I-IV] produces the energy to pump [https://en.wikipedia.org/wiki/Hydrogen_ion hydrogen ions] into the intermembrane space and establish the proton [https://en.wikipedia.org/wiki/Electrochemical_gradient electrochemical gradient] potential.<ref name="Giorgi"/> This negative electrochemical potential is the driving force that moves positively charged calcium ions into the mitochondrial matrix.<ref name="Giorgi"/> Calcium uptake and efflux must be tightly regulated to controll essential [https://en.wikipedia.org/wiki/Citric_acid_cycle Krebs cycle] enzyme activity, including [http://proteopedia.org/wiki/index.php/Pyruvate_dehydrogenase pyruvate dehydrogenase], [https://en.wikipedia.org/wiki/Oxoglutarate_dehydrogenase_complex α-ketoglutarate dehydrogenase], and [http://proteopedia.org/wiki/index.php/Isocitrate_dehydrogenase isocitrate dehydrogenase], while avoiding calcium overload and [https://en.wikipedia.org/wiki/Apoptosis apoptosis].<ref name="Wang"/> | ||
| Line 13: | Line 13: | ||
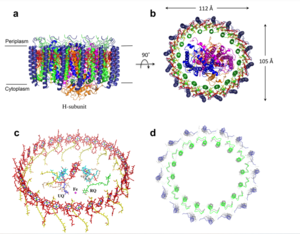
[[Image:structure.png|300 px|right|thumb|Figure 1: Structure of mitochondrial calcium uniporter colored by functional domain. The transmembrane domain is highlighted in salmon, the linker domain spanning the mitochondrial matrix in light cyan, coiled-coil domain in dark violet, and the N-terminal domain in slate blue. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | [[Image:structure.png|300 px|right|thumb|Figure 1: Structure of mitochondrial calcium uniporter colored by functional domain. The transmembrane domain is highlighted in salmon, the linker domain spanning the mitochondrial matrix in light cyan, coiled-coil domain in dark violet, and the N-terminal domain in slate blue. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
==MCU Structure== | ==MCU Structure== | ||
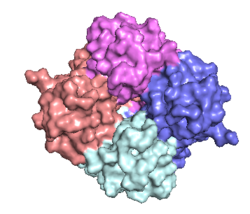
| - | The <scene name='83/832952/Starting_scene/5'>MCU</scene> is the ion channel component of the MCU complex (Figure 1). An NMR structure of an inactive MCU from ''C. elegans'' showed a pentameric arrangement, but the recent crystal and cryo-EM structures of multiple MCUs reaffirmed that active eukaryotic MCU exists as four monomers, identical in sequence, arranged and packed together such that they structurally form a <scene name='83/832952/Dimer_of_dimers/5'>dimer of dimers</scene> in a tetrameric truncated pyramid (Figure 2).<ref name="Woods">PMID:31869674</ref> The MCU protein is composed of a <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene>, a <scene name='83/837230/Coiled_coil/3'>coiled coil domain</scene>, and a <scene name='83/837230/Nterm/2'>N- | + | The <scene name='83/832952/Starting_scene/5'>MCU</scene> is the ion channel component of the MCU complex (Figure 1). An NMR structure of an inactive MCU from ''C. elegans'' showed a pentameric arrangement, but the recent crystal and cryo-EM structures of multiple MCUs reaffirmed that active eukaryotic MCU exists as four monomers, identical in sequence, arranged and packed together such that they structurally form a <scene name='83/832952/Dimer_of_dimers/5'>dimer of dimers</scene> in a tetrameric truncated pyramid (Figure 2).<ref name="Woods">PMID:31869674</ref> The MCU protein is composed of a <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene>, a <scene name='83/837230/Coiled_coil/3'>coiled coil domain</scene>, and a <scene name='83/837230/Nterm/2'>N-terminal domain</scene> (NTD) (Figure 1).<ref name="Woods"/> The hydrophobic <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is located in the ([https://en.wikipedia.org/wiki/Inner_mitochondrial_membrane inner mitochondrial membrane]) while the hydrophilic coiled-coil domain and NTD are positioned in the mitochondrial matrix.<ref name="Baradaran"/> |
[[Image:Nterm.png|250 px|right|thumb|Figure 2: Symmetry and organization of MCU dimer of dimers viewed from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | [[Image:Nterm.png|250 px|right|thumb|Figure 2: Symmetry and organization of MCU dimer of dimers viewed from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
===Transmembrane Domain=== | ===Transmembrane Domain=== | ||
| Line 28: | Line 28: | ||
The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed of multiple layers of acidic amino acids near the narrow mouth of the channel and is responsible for the high affinity and selectivity of the MCU for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp333)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed of multiple layers of acidic amino acids near the narrow mouth of the channel and is responsible for the high affinity and selectivity of the MCU for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp333)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | ||
| - | The <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif consists of <scene name='83/832952/Tryptophan/2'>Trp332</scene> at the N-terminal end, <scene name='83/832952/Selectivity_filter_asp/2'>Asp333</scene>, <scene name='83/832952/Selectivity_filter_glu/3'>Glu336</scene>, and <scene name='83/832952/New_ones/5'>Pro337</scene>.<ref name="Baradaran"/> The negatively charged side chains of Asp333 and <scene name='83/832933/Glu_358/4'>Glu336</scene> point towards the pore.<ref name="Baradaran"/> The <scene name='83/832933/Diameter/2'>diameter</scene> of the pore created by the carboxyl ring on the 4 identical glutamates (Glu336) is about | + | The <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif consists of <scene name='83/832952/Tryptophan/2'>Trp332</scene> at the N-terminal end, <scene name='83/832952/Selectivity_filter_asp/2'>Asp333</scene>, <scene name='83/832952/Selectivity_filter_glu/3'>Glu336</scene>, and <scene name='83/832952/New_ones/5'>Pro337</scene>.<ref name="Baradaran"/> The negatively charged side chains of Asp333 and <scene name='83/832933/Glu_358/4'>Glu336</scene> point towards the pore.<ref name="Baradaran"/> The <scene name='83/832933/Diameter/2'>diameter</scene> of the pore created by the carboxyl ring on the 4 identical glutamates (Glu336) is about 4Å, allowing only a dehydrated Ca<sup>2+</sup> ion to bind. The combination of these radii and high negative charge (Figure 3) account for the selectivity of the MCU. For example, potassium has an [https://en.wikipedia.org/wiki/Ionic_radius ionic radius] of 1.38Å which is much larger than the 1.00Å ionic radius of calcium and thus cannot fit through the pore.<ref name="Baradaran"/> Additionally, even though sodium ions have a similar ionic radius, the +2 charge on calcium is better matched for coordination with the glutamate residues.<ref name="Baradaran"/> |
The additional residues of the WDXXEP motif, <scene name='83/832952/Tryptophan_proline/2'>Trp332 and Pro337</scene> pack against each other, are oriented towards the pore, and serve to stabilize <scene name='83/832952/Selectivity_filter_glu/4'>Asp333 and Glu336</scene>.<ref name="Baradaran"/><ref name="Fan"/> Trp332 stabilizes the carbonyl side chains of Glu336 through <scene name='83/832933/H_bond_trp354_glu358/3'>hydrogen bonding</scene> and anion pi interactions. Approximately one helical turn below the glutamate ring of the selectivity filter, a wider tyrosine ring (12Å) facilitates calcium rehydration after passage through the selectivity pore.<ref name="Fan"/> | The additional residues of the WDXXEP motif, <scene name='83/832952/Tryptophan_proline/2'>Trp332 and Pro337</scene> pack against each other, are oriented towards the pore, and serve to stabilize <scene name='83/832952/Selectivity_filter_glu/4'>Asp333 and Glu336</scene>.<ref name="Baradaran"/><ref name="Fan"/> Trp332 stabilizes the carbonyl side chains of Glu336 through <scene name='83/832933/H_bond_trp354_glu358/3'>hydrogen bonding</scene> and anion pi interactions. Approximately one helical turn below the glutamate ring of the selectivity filter, a wider tyrosine ring (12Å) facilitates calcium rehydration after passage through the selectivity pore.<ref name="Fan"/> | ||
| Line 34: | Line 34: | ||
===Movement of Calcium=== | ===Movement of Calcium=== | ||
| - | Cryo-EM showed three <scene name='83/832952/Starting_scene/5'>calciums</scene> in the MCU channel of roughly spherical density equally spaced 6Å apart.<ref name="Baradaran"/> Sites 1 and 2 lie within the <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> and likely contain calcium, but site 3 could be calcium or some other small molecule.<ref name="Baradaran"/> Site 1 is positioned in the ring formed by <scene name='83/832952/Selectivity_filter_asp/2'>Asp333</scene> residues with a distance of | + | Cryo-EM showed three <scene name='83/832952/Starting_scene/5'>calciums</scene> in the MCU channel of roughly spherical density equally spaced 6Å apart.<ref name="Baradaran"/> Sites 1 and 2 lie within the <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> and likely contain calcium, but site 3 could be calcium or some other small molecule.<ref name="Baradaran"/> Site 1 is positioned in the ring formed by <scene name='83/832952/Selectivity_filter_asp/2'>Asp333</scene> residues with a distance of 8Å between the center of the site and each [https://en.wikipedia.org/wiki/Carboxylate carboxylate group] indicating the presence of water.<ref name="Baradaran"/> Site 2 is positioned in the ring formed by <scene name='83/832952/Selectivity_filter_glu/3'>Glu336</scene> with a smaller distance (4.0Å) between the carboxylate group of each residue and the middle of the site, indicating the absence of water.<ref name="Baradaran"/> For transporting calcium, a mechanism has been proposed where one calcium ion coordinated with water positioned in site 1 is dehydrated and moves to site 2 while a new calcium ion moves from the intermembrane space into site 1.<ref name="Baradaran"/> Meanwhile, a different calcium ion moves from site 2 to site 3 and becomes rehydrated upon passage into the mitochondrial matrix.<ref name="Baradaran"/> |
===Mutations=== | ===Mutations=== | ||
Current revision
Mitochondrial Calcium Uniporter
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0331-8. doi:, 10.1038/s41586-018-0331-8. PMID:29995857 doi:http://dx.doi.org/10.1038/s41586-018-0331-8
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Woods JJ, Wilson JJ. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr Opin Chem Biol. 2019 Dec 20;55:9-18. doi: 10.1016/j.cbpa.2019.11.006. PMID:31869674 doi:http://dx.doi.org/10.1016/j.cbpa.2019.11.006
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 Wang CH, Wei YH. Role of mitochondrial dysfunction and dysregulation of Ca(2+) homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J Biomed Sci. 2017 Sep 7;24(1):70. doi: 10.1186/s12929-017-0375-3. PMID:28882140 doi:http://dx.doi.org/10.1186/s12929-017-0375-3
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
Student Contributors
Ryan Heumann
Lizzy Ratz
Holly Rowe
Madi Summers
Rieser Wells