Sandbox Reserved 1628
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 10: | Line 10: | ||
EpaA1 looks specifically for certain sugars on the outside of the host cells and will bind to those. <scene name='86/861610/Ligandhighlight/2'>This specific adhesin</scene> binds to beta-D-galactopyranose-(1,3)-beta-D-glucopyranose. For ''Candida glabrata'', there are about 20 different types of epithelial adhesins, each with conserved areas of the binding site and variable areas to be able to bind to a variety of sugars on the host cell<ref name="journal">PMID:32669365</ref>. | EpaA1 looks specifically for certain sugars on the outside of the host cells and will bind to those. <scene name='86/861610/Ligandhighlight/2'>This specific adhesin</scene> binds to beta-D-galactopyranose-(1,3)-beta-D-glucopyranose. For ''Candida glabrata'', there are about 20 different types of epithelial adhesins, each with conserved areas of the binding site and variable areas to be able to bind to a variety of sugars on the host cell<ref name="journal">PMID:32669365</ref>. | ||
| - | |||
| - | |||
| - | //The protein is mainly made of antiparallel beta sheets, with the loops connecting the sheets making up the most critical parts of the active site. Two loops responsible for binding to the calcium ion, CBL1 and CLB2, are closer to the rest of the protein, and loops L1-L3 are further out - some of these loops are highly conserved between different adhesins, some are very variable.// | ||
== Biological relevance and broader implications == | == Biological relevance and broader implications == | ||
| Line 25: | Line 22: | ||
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | The majority of the <scene name='86/861610/Secondarystructure/1'>protein's secondary structure</scene> is comprised of antiparallel beta sheets with loops connecting them. Two loops help to form a calcium binding pocket which helps to bind the calcium ion shown and three other loops form the outer pocket. These loops are highly variable in their composition and are contributed with recognizing certain sugars that the inner binding site prefer to bind to. | ||
| + | |||
| + | The tertiary structure allows the loops created by the antiparallel beta sheets <scene name='86/861610/Tertiarystructure/1'>to come close enough to interact with one another</scene>. These loops help to stabilize the calcium ion shown in blue as well as form the outer pocket. Between two of the variable loops is a disulfide bond created by two cystine amino acids which is the only conserved area within the loops. The disulfide bond helps to maintain structure of the outer pocket and is consistent between a variety of the epithelial adhesins. | ||
| + | |||
| + | The protein itself has <scene name='86/861610/Spacefill/2'>a small indent near the active site</scene> where the calcium ion sits just sticking out to interact with sugar ligands. The cleft into the protein fits a very specific size of sugar, which follows along with the current research that shows only a few different pyranose sugars can interact with the adhesins<ref name="journal" />. | ||
== Other important features == | == Other important features == | ||
| - | Tryptophan | + | Although not bound to the ligand, <scene name='86/861610/Tryptophan/1'>tryptophan interacts with it</scene> through hydrophobic interactions and maintains its position within the active site. Changing the amino acid to one without an aromatic ring decreased the affinity for the ligand to bind to the adhesin, while switching it with an amino acid that also has an aromatic ring resulted in no overall change. This shows the most important feature of this specific amino acid - the aromatic ring interacts with the ligand by creating a hydrophobic interaction to stabilize it. |
| + | |||
| + | The calcium ion featured within the active site is not a ligand itself as it's necessary for the protein to work and bind to the sugar ligand. Most of the bonds formed from the ligand are connected to the calcium ion in metal coordination complexes, which in turn is bound to the protein. Two conserved loops within multiple paralogs of the protein are specifically to make sure the calcium ion is within the correct position. | ||
| + | |||
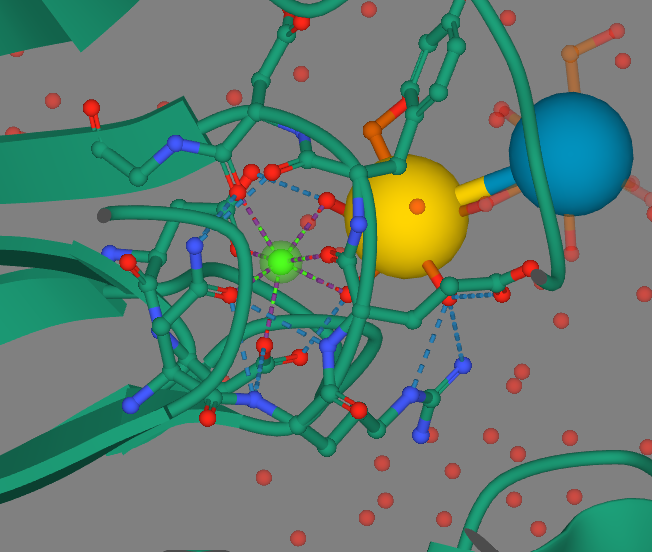
| + | [[Image:CalciumComplex.png]] | ||
| + | |||
| + | Calcium is in green, the yellow and blue spheres represent the sugar ligand, the purple branches out from calcium are metal coordination complexes, and the blue dashed branches are hydrogen bonds. | ||
</StructureSection> | </StructureSection> | ||
Current revision
| This Sandbox is Reserved from 09/18/2020 through 03/20/2021 for use in CHEM 351 Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, IA. This reservation includes Sandbox Reserved 1628 through Sandbox Reserved 1642. |
To get started:
More help: Help:Editing |
Epithelial Adhesin 1A
| |||||||||||
References
- ↑ Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999 Feb;181(4):1059-71. doi: 10.1128/JB.181.4.1059-1071.1999. PMID:9973330 doi:http://dx.doi.org/10.1128/JB.181.4.1059-1071.1999
- ↑ 2.0 2.1 Fidel PL Jr, Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999 Jan;12(1):80-96. PMID:9880475
- ↑ 3.0 3.1 Hoffmann D, Diderrich R, Reithofer V, Friederichs S, Kock M, Essen LO, Mosch HU. Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding. J Biol Chem. 2020 Jul 15. pii: RA120.013968. doi: 10.1074/jbc.RA120.013968. PMID:32669365 doi:http://dx.doi.org/10.1074/jbc.RA120.013968