User:Allison Welz/Sandbox 1
From Proteopedia
< User:Allison Welz(Difference between revisions)
(New page: ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> This is a default text for you...) |
|||
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | = | + | =LPL with GPIHBP-1,'' Drosophilia S2''= |
<StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | + | ||
| - | + | == Introduction == | |
| + | |||
| + | === History === | ||
| + | |||
| + | === Significance === | ||
== Function == | == Function == | ||
| + | |||
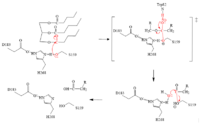
| + | ====Mechanism==== | ||
| + | [[Image:mech426.png|200 px|]] | ||
| + | # The triglyceride binds to LPL’s lipid-binding region in an open lid conformation. | ||
| + | # The oxygen on S159 is made more [https://en.wikipedia.org/wiki/Nucleophile nucleophilic]. This happens via [https://en.wikipedia.org/wiki/Histidine histidine] hydrogen bonding with the hydrogen on S159’s alcohol group. | ||
| + | # The nucleophilic oxygen attacks the [https://en.wikipedia.org/wiki/Carbonyl_group carbonyl carbon] of one of the fatty acid chains. | ||
| + | # This pushes electrons up onto the carbonyl oxygen, creating a [http://www.chem.ucla.edu/~harding/IGOC/T/tetrahedral_intermediate.html tetrahedral intermediate]. This is the oxyanion hole which is stabilized by main chain nitrogen atoms of W82 and L160. | ||
| + | # One of the lone pairs of the oxygen (in the oxyanion hole) creates a double bond carbon. | ||
| + | # The oxygen-carbon bond between the single fatty acid chain and the [https://en.wikipedia.org/wiki/Diglyceride diglyceride] is cleaved. | ||
| + | # H268 hydrogen bonds water, making the oxygen a better nucleophile. Water attacks the carbonyl carbon. | ||
| + | # The [https://en.wikipedia.org/wiki/Carboxylic_acid carboxylic acid] is formed and the S159 bond is cleaved and re-protonated via H268. | ||
| + | # The active site is now back in its original state. | ||
== Disease == | == Disease == | ||
| + | |||
| + | === Mutations === | ||
| + | ====D201V==== | ||
| + | <scene name='87/877636/D201_mutation/10'>D201V</scene> is a mutation that is found to cause [https://en.wikipedia.org/wiki/Lipoprotein_lipase_deficiency chylomicronemia]. Chylomicronemia is when the body cannot break down lipids properly. This leads to their build-up in the body causing high levels of triglycerides in the body. The [https://en.wikipedia.org/wiki/Aspartic_acid carboxyl side chain of aspartate] 201 is one of the coordination sites for the calcium ion of LPL. The mutation to hydrophobic [https://en.wikipedia.org/wiki/Valine valine] means the loss of this coordination site<ref name="Birrane">PMID:30559189</ref>. This mutation adversely affects the folding of LPL and thus affects the secretion of LPL, overall decreasing the activity of LPL<ref name="Birrane">PMID:30559189</ref>. | ||
| + | |||
| + | ====M404R==== | ||
| + | <scene name='87/877636/M404r_1/2'>M404R</scene> is a mutation found within LPL that caused [https://en.wikipedia.org/wiki/Lipoprotein_lipase_deficiency chylomicronemia] in patients. The hydrophobic [https://en.wikipedia.org/wiki/Methionine methionine] is mutated to the larger and charged side chain of [https://en.wikipedia.org/wiki/Arginine arginine]. Originally it was thought to impact LPL secretion from cells. It was found that the M404R does not affect LPL secretion <ref name="Birrane">PMID:30559189</ref>. M404R interacts with the hydrophobic pocket of GPIHBP1’s finger 3 of its 3 fingered domain (V121, E122, T124, V126). The large, charged arginine repelled the hydrophobic pocket and does not fit well. This prevents proper binding and formation of the LPL-GPIHBP1 complex <ref name="Birrane">PMID:30559189</ref>. | ||
| + | </StructureSection> | ||
| + | |||
| + | == M404R Mutation == | ||
| + | <StructureSection load='3rec' size='350' side='right' caption='M404R Mutation w/ GPIHBP-1 Valine Residues' scene='87/877636/M404r_1/1'> | ||
| + | |||
| + | Anything in this section will appear adjacent to the 3D structure and will be scrollable. | ||
| + | |||
| + | </StructureSection> | ||
== Relevance == | == Relevance == | ||
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | == Student Contributors == | ||
| + | |||
| + | *Ashrey Burley | ||
| + | *Allison Welz | ||
| + | *Hannah Wright | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Current revision
Contents |
LPL with GPIHBP-1, Drosophilia S2
| |||||||||||
M404R Mutation
| |||||||||||
Relevance
Structural highlights
</StructureSection>
Student Contributors
- Ashrey Burley
- Allison Welz
- Hannah Wright
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
- ↑ 1.0 1.1 1.2 1.3 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116