This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1678
From Proteopedia
(Difference between revisions)
| (64 intermediate revisions not shown.) | |||
| Line 5: | Line 5: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | == Function == | + | == Function of your protein == |
| + | The protein, <scene name='87/873240/Cartoon_img_of_alyc3_rotating/1'>Alginate lyase (AlyC3)</scene>, is from the Arctic brown alga, Laminara. Alginate lyase is an important enzyme for breaking down alginate in the ocean; alginate is a protein with unique gel-forming properties. The alginate lyase enzyme binds tetramannuronate and polymannuronate as substrates and generates oligosaccharide or monosaccharide products. | ||
| - | + | == Biological relevance and broader implications == | |
| - | + | Studying alginate lyase is crucial to the understanding of how alginate is degraded and recycled in the ocean and to the development of their use for human benefit. Alginate lyases are an important element of the pharmaceutical industry as they can be used to treat chronic pulmonary infections such as cystic fibrosis (CF). The pathogen, Pseudomonas aeruginosa, infects the airway of CF patients and causes excessive lung inflammation, mucus blockage in the airway, and quick declines in overall lung function. Alginate lyase has been shown to participate in detachment of Pseudomonas aeruginosa from bacterial biofilms in the airway. | |
| - | == | + | This particular protein is important to study because little is known about the alginate lyases that originate from cold polar environments. These alginate lyases from the Arctic ocean have some unique mechanisms for adaptation and alginate degradation. |
| + | |||
| + | == Important amino acids== | ||
| + | The amino acids Arg78, Arg82, Gln125, His127 (Ala127 in mutant), Tyr190, and Tyr244 (Ala244 in mutant) are all <scene name='87/873240/Import_amino_acids_substrate/1'>important for binding</scene> the substrate. The catalytic amino acids for alginate lyase (AlyC3) are normally His127 and Tyr244, but this is the mutant alginate lyase, which has <scene name='87/873240/Catalytic_amino_acids/1'>catalytic amino acids</scene> of Ala127 and Ala244. The <scene name='87/873240/Catalytic_triad/1'>catalytic triad</scene>, Arg78, Arg82, and Tyr190, is involved in accurate positioning of the substrate polymer in the catalytic center. | ||
| + | |||
| + | |||
| + | Another view of the protein's active site can be seen <scene name='87/873240/Active_site_amino_acids/1'>here</scene>. The catalytic amino acids (Ala127, Ala244) are shown in blue, catalytic triad (Arg78, Arg82, Tyr190) is shown in purple, and Gln125 (important for hydrogen bonding) is shown in orange. | ||
| + | |||
| + | |||
| + | Alginate lyase contains a main ligand/substrate that is a <scene name='87/873240/7c8f_view_2/1'>tetramannuronate.</scene> The beta pleated sheets, 7 and 10, containing Gln125 and Tyr190, respectively, form <scene name='87/873240/Test_with_dr_hall/5'>important interactions</scene> with the main ligand. | ||
== Structural highlights == | == Structural highlights == | ||
| + | Alginate lyase is a <scene name='87/873240/Dimer/2'>dimer</scene> (contains two domains). One domain is shown in blue, and the other is in green. Being in a dimer form allows the protein to adapt to the seawater salinity of the Arctic ocean from which is originates. The protein's tertiary structure is held together by many hydrogen bonds and some cation-pi interactions. Each domain of the protein is made of <scene name='87/873240/Alpha_helices_and_beta_sheets/1'>4 alpha helices (shown in yellow) and 2 large beta sheets (shown in pink)</scene>. The <scene name='87/873240/Beta_interaction_w_substrate/1'>beta sheets are very important</scene> for interaction with the substrate, mannuronate. One beta sheet is called sheet A, and the other is sheet B. Sheet A consists of 9 beta strands, and sheet B consists of 7 beta strands; you can see the distinction between the two strands <scene name='87/873240/Sheet_a_vs_sheet_b/1'>here</scene> (sheet A = pink, sheet B = orange). <scene name='87/873240/Cleft_in_sheet_a/1'>Sheet A shapes a cleft</scene> and forms a positively charged groove where the substrate binds. | ||
| + | |||
| + | |||
| + | A space-fill view of the protein is shown <scene name='87/873240/Spacefill_cleft_and_substrate/1'>here</scene>, and the cleft in the blue domain can be seen where the substrate binds to the active site. The cleft is a long valley-like groove that is positively charged for the negatively charged long-chained sugar substrate to sit in. The space-fill view also clearly displays the two domains of the protein. | ||
| + | |||
| - | + | == Other important features == | |
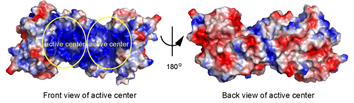
| - | + | As seen in the image below, the protein has two active centers since it is a dimer. In the "front view of the active center," it can be seen that each active center is made of positively charged amino acids (blue) important for binding the negatively charged substrate. As seen in the "back view of the active center," the rest of the protein not containing the active site is evenly distributed with positive and negative charges because the substrate does not bind there. | |
| + | [[Image:AlyC3_active_center_positive_and_negative.png]] | ||
| - | + | AlyC3 is the first dimeric structure of endolytic (cleaving glycosidic bonds in the middle) alginate lyases. As mentioned above, the dimeric structure allows the protein to adapt to the salinity of the seawater where it lives. <scene name='87/873240/Dimer_loop_2/1'>Loop 2</scene> (yellow) of AlyC3 is essential to maintaining the dimeric structure; a disulfide bond between Cys170 and Cys184 cross-links loop 2. | |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | <ref> PMID 32967968 </ref> | ||
| + | <ref> PMID 23070175 </ref> | ||
<references/> | <references/> | ||
| - | <ref> 32967968 </ref> | ||
Current revision
| This Sandbox is Reserved from 01/25/2021 through 04/30/2021 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1665 through Sandbox Reserved 1682. |
To get started:
More help: Help:Editing |
Alginate Lyase (AlyC3)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Xu F, Chen XL, Sun XH, Dong F, Li CY, Li PY, Ding H, Chen Y, Zhang YZ, Wang P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the Arctic. J Biol Chem. 2020 Sep 23. pii: RA120.015106. doi: 10.1074/jbc.RA120.015106. PMID:32967968 doi:http://dx.doi.org/10.1074/jbc.RA120.015106
- ↑ Lamppa JW, Griswold KE. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob Agents Chemother. 2013 Jan;57(1):137-45. doi: 10.1128/AAC.01789-12., Epub 2012 Oct 15. PMID:23070175 doi:http://dx.doi.org/10.1128/AAC.01789-12