User:Josey McKinley/Sandbox 1
From Proteopedia
< User:Josey McKinley(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 6: | Line 6: | ||
== Introduction== | == Introduction== | ||
| - | |||
[http://en.wikipedia.org/wiki/Stearoyl-CoA_desaturase-1 Stearoyl-CoA Desaturase 1 (SCD1)] is an iron-containing [http://en.wikipedia.org/wiki/Fatty_acid_desaturase Δ-9-desaturase] that is a key regulator of fatty-acid metabolism where it catalyzes the rate-limiting step in the conversion of [http://en.wikipedia.org/wiki/Stearoyl-CoA Stearoyl-CoA] to [http://en.wikipedia.org/wiki/Oleic_acid oleic acid], an essential substrate in the biosynthesis of phospholipids, triacyclglycerols, and cholesterol. SCD1 is embedded within the membrane of the [http://micro.magnet.fsu.edu/cells/endoplasmicreticulum/endoplasmicreticulum.html endoplasmic reticulum] and consists of 4 transmembrane alpha helices and 11 cytosolic helices. | [http://en.wikipedia.org/wiki/Stearoyl-CoA_desaturase-1 Stearoyl-CoA Desaturase 1 (SCD1)] is an iron-containing [http://en.wikipedia.org/wiki/Fatty_acid_desaturase Δ-9-desaturase] that is a key regulator of fatty-acid metabolism where it catalyzes the rate-limiting step in the conversion of [http://en.wikipedia.org/wiki/Stearoyl-CoA Stearoyl-CoA] to [http://en.wikipedia.org/wiki/Oleic_acid oleic acid], an essential substrate in the biosynthesis of phospholipids, triacyclglycerols, and cholesterol. SCD1 is embedded within the membrane of the [http://micro.magnet.fsu.edu/cells/endoplasmicreticulum/endoplasmicreticulum.html endoplasmic reticulum] and consists of 4 transmembrane alpha helices and 11 cytosolic helices. | ||
| Line 15: | Line 14: | ||
SCD stands for Stearoyl-CoA Desaturase. This enzyme is highly conserved in eukaryotes and has different isoforms. Mice have four isoforms: SCD1, SCD2, SCD3, and SCD4. Humans have two different isoforms: SCD1 and SCD5. The SCD discussed in this page is the SCD-1 found in mice. SCD was thought to have once been an anaerobic pathway found in cartilaginous fish about 450 million years ago. The enzyme’s mechanism is now aerobic and this aerobic pathway is favored. The structure of SCD1 was found using X-ray crystallography. This enzyme is embedded in the membrane of the Endoplasmic Reticulum. SCD is made up of 15 alpha helices; four helices are embedded in the membrane and 11 helices in the cytosol. The substrate binds to the helices in the cytosol. | SCD stands for Stearoyl-CoA Desaturase. This enzyme is highly conserved in eukaryotes and has different isoforms. Mice have four isoforms: SCD1, SCD2, SCD3, and SCD4. Humans have two different isoforms: SCD1 and SCD5. The SCD discussed in this page is the SCD-1 found in mice. SCD was thought to have once been an anaerobic pathway found in cartilaginous fish about 450 million years ago. The enzyme’s mechanism is now aerobic and this aerobic pathway is favored. The structure of SCD1 was found using X-ray crystallography. This enzyme is embedded in the membrane of the Endoplasmic Reticulum. SCD is made up of 15 alpha helices; four helices are embedded in the membrane and 11 helices in the cytosol. The substrate binds to the helices in the cytosol. | ||
| + | |||
| + | ==Function== | ||
==Binding of the Substrate== | ==Binding of the Substrate== | ||
Stearoyl-CoA is the substrate that binds to the enzyme, SCD1. The binding of the substrate is stabilized by specific residues on the exterior and interior of the protein. Stearoyl-CoA is a long-chain fatty acyl-CoA. The head group of the substrate is composed of an adenine, ribose, phosphate groups, and many atoms of Nitrogen, oxygen, and sulfur. The fatty acid tail is a 17-carbon chain which reaches into the interior of the protein. The head of stearoyl-CoA is attached to the exterior of the protein by polar residues. The adenine, ribose, and phosphate are attached via <scene name='87/877602/Hydrophillic_top/2'>Hydrophillic Top</scene> groups that include R151, D152, and K185. The rest of the exterior of the substrate is attached via <scene name='87/877602/Hydrophillic_bottom_labeled/2'>Hydrophillic Bottom</scene> groups that include R184, N144, and N71. The fatty acid chain dives into the interior of the enzyme. The acyl chain is enclosed in a tunnel (24 ang) buried in the cytosolic domain. The geometry of the tunnel and formation of bound acyl chain are the structural basis for the stereospecificity of the desaturation reaction. The chain is kinked at carbon 9-carbon 10 where the double bond is generated. The <scene name='87/877602/Full_kink/2'>Kink</scene> is induced through the interactions of four conserved residues. Three out of four of these residues are not bound to the chain, but are hydrogen bonded to each other: <scene name='87/877602/Kink/2'>W149, T257, and Q143</scene>. T257 is hydrogen bonded to Q143, and Q143 is hydrogen bonded to W149. These residues are directly below the kink and will be hydrolyzed when the substrate is ready to be released. The residue that is directly hydrogen bonded to the chain is <scene name='87/877602/Trp258/2'>Trp258</scene>. This residue is highly conserved and stabilizes the chain so it will be in the correct orientation in the active site. | Stearoyl-CoA is the substrate that binds to the enzyme, SCD1. The binding of the substrate is stabilized by specific residues on the exterior and interior of the protein. Stearoyl-CoA is a long-chain fatty acyl-CoA. The head group of the substrate is composed of an adenine, ribose, phosphate groups, and many atoms of Nitrogen, oxygen, and sulfur. The fatty acid tail is a 17-carbon chain which reaches into the interior of the protein. The head of stearoyl-CoA is attached to the exterior of the protein by polar residues. The adenine, ribose, and phosphate are attached via <scene name='87/877602/Hydrophillic_top/2'>Hydrophillic Top</scene> groups that include R151, D152, and K185. The rest of the exterior of the substrate is attached via <scene name='87/877602/Hydrophillic_bottom_labeled/2'>Hydrophillic Bottom</scene> groups that include R184, N144, and N71. The fatty acid chain dives into the interior of the enzyme. The acyl chain is enclosed in a tunnel (24 ang) buried in the cytosolic domain. The geometry of the tunnel and formation of bound acyl chain are the structural basis for the stereospecificity of the desaturation reaction. The chain is kinked at carbon 9-carbon 10 where the double bond is generated. The <scene name='87/877602/Full_kink/2'>Kink</scene> is induced through the interactions of four conserved residues. Three out of four of these residues are not bound to the chain, but are hydrogen bonded to each other: <scene name='87/877602/Kink/2'>W149, T257, and Q143</scene>. T257 is hydrogen bonded to Q143, and Q143 is hydrogen bonded to W149. These residues are directly below the kink and will be hydrolyzed when the substrate is ready to be released. The residue that is directly hydrogen bonded to the chain is <scene name='87/877602/Trp258/2'>Trp258</scene>. This residue is highly conserved and stabilizes the chain so it will be in the correct orientation in the active site. | ||
| - | = | + | <scene name='87/877627/Just_zn_but_zoomed_out/1'>TextToBeDisplayed</scene> |
| + | == Active Site== | ||
| + | |||
| + | Using X-ray crystallography, two structures of SCD have been found, differing only in their dimetal center. One structure includes the substrate [https://en.wikipedia.org/wiki/Stearoyl-CoA (Stearoyl CoA)] a water molecule, and two <scene name='87/877627/Just_zn_but_zoomed_out/4'>zinc </scene> ions in the center [https://www.rcsb.org/structure/4YMK (4YMK)] (Yonghong). A second structure found more recently includes the product [https://en.wikipedia.org/wiki/Oleic_acid (Oleic Acid)] and two <scene name='87/877627/Zoomed_out_fe/4'>iron</scene> ions in the center [https://www.rcsb.org/structure/6WF2 (6WF2)]. When testing the Zn-centered structure, the enzyme was found to be inactive (Shen). The Zn ions serve as a surrogate for Fe as Zn did not display its typical coordination geometry, tetrahedral; instead, it displayed octahedral geometry which is typical of Fe ion coordination (Yonghong). For the images and links below, the zinc ion-centered structure will be used as it includes the substrate, even though iron is confirmed as the dimetal center. | ||
| + | |||
| + | The dimetal center is essential to the catalytic activity, as previously demonstrated in the mechanism above. The <scene name='87/877627/Zn_with_measurement/3'>zinc</scene> ions are 6.4 angstroms apart (Yonghong). The ions sit above the kink created by C9 and C10 of the substrate within the active site. The ions are held into the active site through the <scene name='87/877627/His_box_w_o_water/3'>His box</scene> (Kohtaro). The nine coordinating His residues stabilize the ions into the active site forming a non-heme prosthetic group (Kohtaro). The His box is highly conserved among the isoforms of SCD (Shen). | ||
| + | |||
| + | The <scene name='87/877627/Zn2/2'>ion</scene> closest to C10 of the substrate is 4.7 angstroms away from this carbon (Yonghong). This ion is coordinated by five histidine residues. The <scene name='87/877627/Zn1/3'>ion</scene> closest to C9 of the substrate is 5.2 angstroms away from this carbon (Yonghong). This ion is coordinated with four histidine residues and one water molecule. The <scene name='87/877627/Zn_and_water_round_2/5'>water</scene> is in coordination to the zinc ion closest to it. It occupies the fifth <scene name='87/877627/His_box_w_o_labels/3'>coordination site</scene>. | ||
| + | |||
| + | Residues around the periphery hydrogen bond to the His box to stabilize it. These residues include <scene name='87/877627/D165_correct_one/5'>D165</scene> <scene name='87/877627/E291_correct_one/3'>E291</scene> and <scene name='87/877627/E161_correct_one/3'>E161</scene> (Yonghong). Another residue that stabilizes the active site is <scene name='87/877627/N261_correct_one/4'>N261</scene>. This residue hydrogen bonds to the water molecule (Yonghong). | ||
| + | The His box and periphery residues stabilize the dimetal center and make up the <scene name='87/877627/Active_site_round_3_but_labels/3'>active site</scene> of the enzyme. This allows for the proposed above mechanism to be carried out. | ||
| + | |||
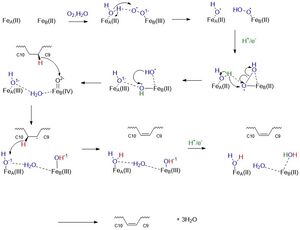
| + | ==Mechanism== | ||
| + | [[Image:SCDMech.jpg|300px|thumb|left|]] | ||
== Disease == | == Disease == | ||
| - | + | Biological relevance | |
| - | + | ||
| - | + | SCD1 role in converting stearoyl CoA, a saturated fatty acid, to oleic acid, a monounsaturated fatty acid, is essential in lipid metabolism. Fluxes in the ratio of saturated fatty acids to monounsaturated fatty acids can be connected to many different disease states, including obesity, diabetes, cancer, and cardiovascular disease (Ntambi). | |
| - | == | + | The inactivation of SCD1 has been known to have combative effects on obesity and diabetes. Increased levels of oleic acid are present in both obesity and diabetes; therefore, inactivating the enzyme will allow for decreased amounts of product present (Ahmed). The inactivation of SCD1 is most commonly seen as a mutation to any of the nine histidine residues present in the <scene name='87/877627/His_box_w_o_water/3'>His box</scene> (Yonghong). A mutation in any of these positions leads to a nonfunctional enzyme. The inactivation of SCD1 also has been known to inhibit cancer cell growth (Shen). |
| + | The inactivation of SCD1 is also commonly caused by the insertion of a proline at position 279. In the wild type SCD1 protein, this position contains an <scene name='87/877627/R279/4'>arginine residue</scene>. A ‘CCC’ codon is inserted into the 835th position of exon 5 in the SCD1 gene. This mutation results in a loss of function of SCD1. This study was done using a mouse model. In mice with this mutation, hair loss, similar to alopecia, occurs. The mice were also found to be lean throughout their lifespan due to reduced triglyceride synthesis connected to the loss of SCD1 function (Y Lu). | ||
| - | <scene name='87/877627/Zn_and_water/1'>Zn and HOH</scene> | ||
| - | <scene name='87/877627/Just_zn/1'>Zn ions</scene> | ||
| - | <scene name='87/877627/His_box2/1'>His box</scene> | ||
| - | <scene name='87/877627/E161/1'>E161</scene> | ||
| - | <scene name='87/877627/N261/1'>N261</scene> | ||
| - | <scene name='87/877627/D165/1'>D165</scene> | ||
| - | <scene name='87/877627/E291/1'>E291</scene> | ||
| - | <scene name='87/877627/As_bigboy_zngray/1'>overall active site</scene> | ||
| - | <scene name='87/877602/Full_kink/2'>Full Kink</scene> | ||
| - | <scene name='87/877602/Trp258/2'>Trp258</scene> | ||
| - | <scene name='87/877602/Hydrophillic_bottom_labeled/2'>Hydrophilic Bottom</scene> | ||
| - | <scene name='87/877602/Hydrophillic_top/2'>Hydrophilic Top</scene> | ||
| - | <scene name='87/877602/Kink/2'>Kink</scene> | ||
| - | <scene name='87/877602/Hydrophillic_bottom_labeled/1'>Hydrophilic interactions</scene> | ||
</StructureSection> | </StructureSection> | ||
| - | <scene name='87/877602/Hydrophillic_top/1'>Abbey's Scene</scene> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
| - | <ref name=" | + | <ref name="Bai">PMID:26098370</ref> |
| + | <ref name="Shen">PMID:32470559</ref> | ||
| + | <ref name="Filipe">doi: 10.1186/1471-2148-11-132</ref> | ||
| + | <ref name="Paton">doi: 10.1152/ajpendo.90897.2008</ref> | ||
| + | <ref name="Lu">PMID: 15278437</ref> | ||
| + | <ref name="ALJohani">PMID: 29089222</ref> | ||
| + | <ref name="Kikuchi">PMID: 31838050</ref> | ||
| + | <ref name="Ntambi">PMID: 14654089</ref> | ||
| + | |||
<references/> | <references/> | ||
Current revision
Stearoyl-CoA Desaturase 1 from Mus musculus
| |||||||||||
References
[3] [4] [5] [6] [7] [8] [9] [10]
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Bai Y, McCoy JG, Levin EJ, Sobrado P, Rajashankar KR, Fox BG, Zhou M. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 2015 Jun 22. doi: 10.1038/nature14549. PMID:26098370 doi:http://dx.doi.org/10.1038/nature14549
- ↑ Shen J, Wu G, Tsai AL, Zhou M. Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase. J Mol Biol. 2020 May 27. pii: S0022-2836(20)30367-3. doi:, 10.1016/j.jmb.2020.05.017. PMID:32470559 doi:http://dx.doi.org/10.1016/j.jmb.2020.05.017
- ↑ Castro LF, Wilson JM, Goncalves O, Galante-Oliveira S, Rocha E, Cunha I. The evolutionary history of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol Biol. 2011 May 19;11:132. doi: 10.1186/1471-2148-11-132. PMID:21595943 doi:http://dx.doi.org/10.1186/1471-2148-11-132
- ↑ Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009 Jul;297(1):E28-37. doi:, 10.1152/ajpendo.90897.2008. Epub 2008 Dec 9. PMID:19066317 doi:http://dx.doi.org/10.1152/ajpendo.90897.2008
- ↑ Lu Y, Bu L, Zhou S, Jin M, Sundberg JP, Jiang H, Qian M, Shi Y, Zhao G, Kong X, Hu L. Scd1ab-Xyk: a new asebia allele characterized by a CCC trinucleotide insertion in exon 5 of the stearoyl-CoA desaturase 1 gene in mouse. Mol Genet Genomics. 2004 Sep;272(2):129-37. doi: 10.1007/s00438-004-1043-3. Epub , 2004 Jul 29. PMID:15278437 doi:http://dx.doi.org/10.1007/s00438-004-1043-3
- ↑ ALJohani AM, Syed DN, Ntambi JM. Insights into Stearoyl-CoA Desaturase-1 Regulation of Systemic Metabolism. Trends Endocrinol Metab. 2017 Dec;28(12):831-842. doi: 10.1016/j.tem.2017.10.003., Epub 2017 Oct 28. PMID:29089222 doi:http://dx.doi.org/10.1016/j.tem.2017.10.003
- ↑ Kikuchi K, Tsukamoto H. Stearoyl-CoA desaturase and tumorigenesis. Chem Biol Interact. 2020 Jan 25;316:108917. doi: 10.1016/j.cbi.2019.108917. Epub , 2019 Dec 12. PMID:31838050 doi:http://dx.doi.org/10.1016/j.cbi.2019.108917
- ↑ Ntambi JM, Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog Lipid Res. 2004 Mar;43(2):91-104. doi: 10.1016/s0163-7827(03)00039-0. PMID:14654089 doi:http://dx.doi.org/10.1016/s0163-7827(03)00039-0
Student Contributors
- Josey McKinley
- Abbey Wells
- Anthony Durand