Sandbox Reserved 1678
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 30: | Line 30: | ||
== Other important features == | == Other important features == | ||
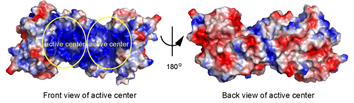
| - | As seen in the image below, the protein has two active centers since it is a dimer. In the "front view of the active center," it can be seen that each active center is made of positively charged amino acids (blue) important for binding the negatively charged substrate. | + | As seen in the image below, the protein has two active centers since it is a dimer. In the "front view of the active center," it can be seen that each active center is made of positively charged amino acids (blue) important for binding the negatively charged substrate. As seen in the "back view of the active center," the rest of the protein not containing the active site is evenly distributed with positive and negative charges because the substrate does not bind there. |
| + | |||
[[Image:AlyC3_active_center_positive_and_negative.png]] | [[Image:AlyC3_active_center_positive_and_negative.png]] | ||
| - | + | AlyC3 is the first dimeric structure of endolytic (cleaving glycosidic bonds in the middle) alginate lyases. As mentioned above, the dimeric structure allows the protein to adapt to the salinity of the seawater where it lives. <scene name='87/873240/Dimer_loop_2/1'>Loop 2</scene> (yellow) of AlyC3 is essential to maintaining the dimeric structure; a disulfide bond between Cys170 and Cys184 cross-links loop 2. | |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | <ref> PMID 32967968 </ref> | ||
| + | <ref> PMID 23070175 </ref> | ||
<references/> | <references/> | ||
| - | <ref> 32967968 </ref> | ||
| - | <ref> 23070175 </ref> | ||
Current revision
| This Sandbox is Reserved from 01/25/2021 through 04/30/2021 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1665 through Sandbox Reserved 1682. |
To get started:
More help: Help:Editing |
Alginate Lyase (AlyC3)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Xu F, Chen XL, Sun XH, Dong F, Li CY, Li PY, Ding H, Chen Y, Zhang YZ, Wang P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the Arctic. J Biol Chem. 2020 Sep 23. pii: RA120.015106. doi: 10.1074/jbc.RA120.015106. PMID:32967968 doi:http://dx.doi.org/10.1074/jbc.RA120.015106
- ↑ Lamppa JW, Griswold KE. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob Agents Chemother. 2013 Jan;57(1):137-45. doi: 10.1128/AAC.01789-12., Epub 2012 Oct 15. PMID:23070175 doi:http://dx.doi.org/10.1128/AAC.01789-12