We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sarah Maarouf/Sandbox 1
From Proteopedia
< User:Sarah Maarouf(Difference between revisions)
| (31 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
== Introduction == | == Introduction == | ||
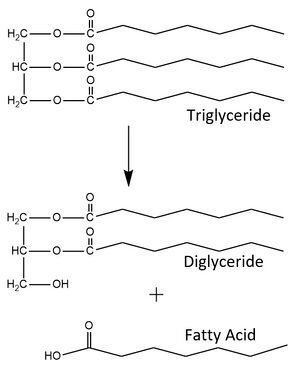
| - | [[Image: | + | [[Image: BasicMech2.jpg|300px|right|thumb|Figure 1. LPL catalyzes the breakdown of triglycerides into a diglyceride and a fatty acid. It can also recognize the diglyceride as a substrate and produce a monoglyceride and two fatty acids from the triglyceride substrate.]] |
| - | <scene name='87/877554/Lpl_w_gpihbp1/4'>Lipoprotein lipase (LPL)</scene> is an enzyme synthesized and secreted primarily by [https://en.wikipedia.org/wiki/Myocyte myocytes] and [https://en.wikipedia.org/wiki/Adipocyte adipocytes] into interstitial spaces.<ref name = "Fong">PMID: 27185325</ref> It is located on the surface of capillaries where it is bound to a glycolipid-anchored protein expressed by capillary endothelial cells. This protein is called glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1, or [https://en.wikipedia.org/wiki/GPIHBP1 GPIHBP1].<ref name="Voss">PMID:21518912</ref> | + | <scene name='87/877554/Lpl_w_gpihbp1/4'>Lipoprotein lipase (LPL)</scene> is an enzyme synthesized and secreted primarily by [https://en.wikipedia.org/wiki/Myocyte myocytes] and [https://en.wikipedia.org/wiki/Adipocyte adipocytes] into interstitial spaces.<ref name = "Fong">PMID: 27185325</ref> It is located on the surface of capillaries where it is bound to a glycolipid-anchored protein expressed by capillary endothelial cells. This protein is called glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1, or [https://en.wikipedia.org/wiki/GPIHBP1 GPIHBP1].<ref name="Voss">PMID:21518912</ref> LPL is an essential enzyme for triglyceride metabolism and utilization, however it is susceptible to unfolding in its catalytic domain, and thus must be bound to GPIHBP1 to prevent loss of enzymatic activity. When LPL is not bound to GPIHBP1 its enzymatic activity is relatively low and declines until it has lost all function. However, when bound to GPIHBP1 it is able to maintain its maximum enzymatic activity.<ref name="Arora">PMID:31072929</ref> In addition, binding to GPIHBP1 is required for adhesion of [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/triglyceride-rich-lipoprotein#:~:text=Triglyceride%2Drich%20lipoproteins%20are%20secreted,phospholipid%20monolayer%20with%20associated%20apolipoproteins triglyceride rich lipoproteins (TRLs)] to LPL and transport of LPL to its site of action in the capillary lumen. Once it has reached the site of action, the enzyme is able to produce a monoglyceride and two fatty acids from the triglyceride substrate (Figure 1).<ref name="Young">PMID:31269429</ref> |
| Line 27: | Line 27: | ||
== Structure == | == Structure == | ||
| - | The [https://proteopedia.org/wiki/index.php/Asymmetric_Unit asymmetric unit] is a <scene name='87/877554/Asymmetric_unit/3'>tetramer</scene> of LPL | + | The [https://proteopedia.org/wiki/index.php/Asymmetric_Unit asymmetric unit] is a <scene name='87/877554/Asymmetric_unit/3'>tetramer</scene> of LPL-GPIHBP1 complexes. The orientation of these four dimers are in a head-to-tail conformation. Despite the unique orientation of each of the four dimers within the tetramer, all act independently and perform the same enzymatic function. Each complex consists of the LPL with N-terminal and C-terminal domains, and the associated GPIHBP1.<ref name="Arora">PMID:31072929</ref> |
=== N-terminal α/β-hydrolase domain of LPL === | === N-terminal α/β-hydrolase domain of LPL === | ||
| - | The <scene name='87/877554/N-terminal_domain/4'>N-terminal domain</scene> is composed of one antiparallel β-sheet and seven parallel β-sheets that are located between five α-helices. This is the domain responsible for hydrolysis of lipid substrates, as it contains the <scene name='87/877554/Active_site_residues/11'>catalytic triad</scene> and houses the <scene name='87/877554/Oxyanion_hole/10'>oxyanion hole</scene> to stabilize the transition state of the substrate. The N-terminal domain includes a <scene name='87/877554/Calcium_ion_coordination_sites/6'>calcium ion that is coordinated by a number of residues</scene> which has been shown to have mutations that may impact LPL enzyme activity. The lid region of the N-terminal domain was imaged in an open conformation, meaning it is not blocking the active site. The <scene name='87/877554/Lid_region/7'>lid region</scene> consists of 2 short α-helices connected by a loop, extending away from the protein. This open conformation allows for many surface-exposed hydrophobic residues (valines,isoleucines, and leucines) to create a hydrophobic patch on the surface of LPL. The lid region helps to control for the entry of lipid substrates into the active site cleft, though specifics about how lipids enter into the active site still | + | The <scene name='87/877554/N-terminal_domain/4'>N-terminal domain</scene> is composed of one antiparallel β-sheet and seven parallel β-sheets that are located between five α-helices. This is the domain responsible for hydrolysis of lipid substrates, as it contains the <scene name='87/877554/Active_site_residues/11'>catalytic triad</scene> and houses the <scene name='87/877554/Oxyanion_hole/10'>oxyanion hole</scene> to stabilize the transition state of the substrate. The N-terminal domain includes a <scene name='87/877554/Calcium_ion_coordination_sites/6'>calcium ion that is coordinated by a number of residues</scene> which has been shown to have mutations that may impact LPL enzyme activity. The lid region of the N-terminal domain was imaged in an open conformation, meaning it is not blocking the active site. The <scene name='87/877554/Lid_region/7'>lid region</scene> consists of 2 short α-helices connected by a loop, extending away from the protein. This open conformation allows for many surface-exposed hydrophobic residues (valines, isoleucines, and leucines) to create a hydrophobic patch on the surface of LPL. The lid region helps to control for the entry of lipid substrates into the active site cleft, though specifics about how lipids enter into the active site still requires further investigation.<ref name="Arora">PMID:31072929</ref> |
=== C-terminal β-barrel domain of LPL === | === C-terminal β-barrel domain of LPL === | ||
| - | The <scene name='87/877554/C-terminal_domain/8'>C-terminal domain</scene> of LPL is made up of 8 antiparallel β-sheets.<ref name="Arora">PMID:31072929</ref> It includes the GPIHBP1 binding site and the <scene name='87/877554/Lipid_binding_region/6'>tryptophan-rich lipid binding region</scene> that helps to contribute to the specificity of this enzyme for TRLs by creating a second hydrophobic patch on the same face of LPL as the active site. The hydrophobic patches are believed to allow for the enzyme to bind TRL substrates with an orientation that facilitates delivery of triglycerides into the active site for catalysis. <ref name="Birrane">PMID:30559189</ref> The lid region and the lipid binding region mechanism of interaction and protein flexibility for binding TRL substrates still | + | The <scene name='87/877554/C-terminal_domain/8'>C-terminal domain</scene> of LPL is made up of 8 antiparallel β-sheets.<ref name="Arora">PMID:31072929</ref> It includes the GPIHBP1 binding site and the <scene name='87/877554/Lipid_binding_region/6'>tryptophan-rich lipid binding region</scene> that helps to contribute to the specificity of this enzyme for TRLs by creating a second hydrophobic patch on the same face of LPL as the active site and the lid region. The hydrophobic patches are believed to allow for the enzyme to bind TRL substrates with an orientation that facilitates delivery of triglycerides into the active site for catalysis. <ref name="Birrane">PMID:30559189</ref> The lid region and the lipid binding region mechanism of interaction and protein flexibility for binding TRL substrates still needs further investigation. <ref name="Arora">PMID:31072929</ref> |
=== GPIHBP1 === | === GPIHBP1 === | ||
[[Image: Electro.jpg|250px|right|thumb|Figure 2. A cartoon representation of GPIHBP1's N-terminal intrinsically disordered region (IDR) colored green and an surface representation of LPL with electrostatic coloring [acidic (red), neutral (white), basic (blue)] to show how GPIHBP1's N-terminal domain interacts with LPL's basic patch to stabilize LPL structure and activity.]] | [[Image: Electro.jpg|250px|right|thumb|Figure 2. A cartoon representation of GPIHBP1's N-terminal intrinsically disordered region (IDR) colored green and an surface representation of LPL with electrostatic coloring [acidic (red), neutral (white), basic (blue)] to show how GPIHBP1's N-terminal domain interacts with LPL's basic patch to stabilize LPL structure and activity.]] | ||
| - | GPIHBP1 is a membrane bound protein that binds to the C-terminal domain of LPL. It contains specific features, the cysteine-rich LU domain and an intrinsically disordered acidic N-terminal domain, that contribute to its binding affinity of LPL. GPIHBP1’s [https://en.wikipedia.org/wiki/LU_domain#:~:text=The%20LU%20domain%20(Ly%2D6,CD59%2C%20and%20Sgp%2D2. LU domain (Ly6-uPar)] is 75 residues in length and adopts a <scene name='87/877554/Fingers_of_gpihbp1/3'>three-fingered fold</scene>, characteristic of the LU protein family, that is stabilized by <scene name='87/877554/Disulfide_bonds_gpihbp1/6'>five disulfide bonds</scene>. The LPL-GPIHBP1 binding interface involves the three-fingered fold and depends largely on hydrophobic interactions between the two subunits | + | GPIHBP1 is a membrane bound protein that binds to the C-terminal domain of LPL. It contains specific features, such as the cysteine-rich LU domain and an intrinsically disordered acidic N-terminal domain, that contribute to its binding affinity of LPL. GPIHBP1’s [https://en.wikipedia.org/wiki/LU_domain#:~:text=The%20LU%20domain%20(Ly%2D6,CD59%2C%20and%20Sgp%2D2. LU domain (Ly6-uPar)] is 75 residues in length and adopts a <scene name='87/877554/Fingers_of_gpihbp1/3'>three-fingered fold</scene>, a characteristic of the LU protein family, that is stabilized by <scene name='87/877554/Disulfide_bonds_gpihbp1/6'>five disulfide bonds</scene>. The LPL-GPIHBP1 binding interface involves the three-fingered fold and depends largely on hydrophobic interactions between the two subunits which creates a high binding affinity of LPL. The disordered acidic N-terminal domain of GPIHBP1 plays a role in capturing LPL, also contributing to binding affinity.<ref name = "Fong">PMID: 27185325</ref> The disordered N-terminal domain has 21 of the 26 total residues being aspartates or glutamates. Unfortunately images for the acidic domain were not resolved, indicating that it likely exhibits conformational flexibility.<ref name="Arora">PMID:31072929</ref> Because GPIHBP1 makes electrostatic interactions with LPL's basic patch (Figure 2), this complex between GPIHBP1's acidic N-terminal domain and LPL's C-terminal domain is speculated to create the stability in LPL that allows for its catalytic activity to continue when bound to GPIHBP1.<ref name="Birrane">PMID:30559189</ref> |
| Line 56: | Line 56: | ||
== Active Site == | == Active Site == | ||
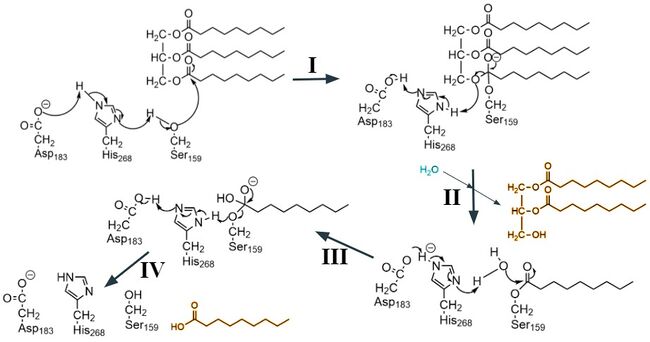
| - | <scene name='87/877554/Active_site_residues/11'>The active site</scene> by which this enzyme catalyzes the hydrolysis of triglycerides | + | <scene name='87/877554/Active_site_residues/11'>The active site</scene> by which this enzyme catalyzes the hydrolysis of triglycerides is located in the N-terminal domain of LPL, and the catalytic residues involved in this reaction are Ser159, Asp183, and His268.<ref name="Birrane">PMID:30559189</ref> Located near these residues, the <scene name='87/877554/Oxyanion_hole/10'>oxyanion hole</scene> stabilizes the transition state of the substrate through the backbone amides of Trp82 and Leu160.<ref name="Arora">PMID:31072929</ref> Surrounding the active site is the <scene name='87/877554/Hydrophobic_binding_region/6'> hydrophobic binding region </scene> which stabilizes the hydrophobic lipid substrates in the binding pocket and forms Van der Waals interactions with the hydrophobic tails of the lipids that are entering. This hydrophobic region also allows the substrates to access the <scene name='87/877554/Access_to_ct/6'>catalytic triad</scene> for hydrolysis.<ref name="Birrane">PMID:30559189</ref> |
=== Mechanism === | === Mechanism === | ||
| Line 94: | Line 94: | ||
| - | The active site consists of a catalytic triad Ser159, Asp183 and His268 that go through the [https://en.wikipedia.org/wiki/Serine_protease#Chymotrypsin-like serine protease mechanism of action]. The hallmark of this mechanism is the proton shuttle between the three residues that increases the nucleophilicity of the serine residue. Serine is then able to make the nucleophilic attack on the the carbonyl carbon of the scissile peptide bond of the substrate. | + | |
| + | The active site consists of a catalytic triad Ser159, Asp183 and His268 that go through the [https://en.wikipedia.org/wiki/Serine_protease#Chymotrypsin-like serine protease mechanism of action]. The hallmark of this mechanism is the proton shuttle between the three residues that increases the nucleophilicity of the serine residue. Serine is then able to make the nucleophilic attack on the the carbonyl carbon of the scissile peptide bond of the substrate (step I). There are several intermediates generated in this ordered mechanism. The first round of the catalysis is where the triglyceride binds, and the diglyceride is released (step II). Then the second substrate, water, binds (step III) and the second product, the fatty acid, is released (step IV).<ref name="Hedstrom">Hedstrom L. Serine Protease Mechanism and Specificity. Chemical Reviews 2002 102 (12), 4501-4524 https://doi.org/10.1021/cr000033x</ref> | ||
| Line 101: | Line 102: | ||
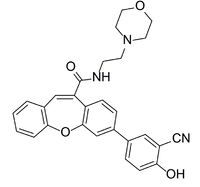
[[Image: Compound_2.jpg|200px|left|thumb|Figure 4. The structure of the inhibitor referred to as Compound 2.]] | [[Image: Compound_2.jpg|200px|left|thumb|Figure 4. The structure of the inhibitor referred to as Compound 2.]] | ||
| - | The active site residues of LPL can be blocked from preforming the breakdown of triglycerides by the presence of a competitive <scene name='87/877554/Compound_2/ | + | The active site residues of LPL can be blocked from preforming the breakdown of triglycerides by the presence of a competitive <scene name='87/877554/Compound_2/4'>inhibitor</scene>. The inhibitor binds in the hydrophobic binding region and occludes access to the catalytic triad. The inhibitor known as Compound 2 (Figure 4) was designed to stabilize the structure of the LPL-GPIHBP1 complex for the purpose of structure determination.<ref name="Arora">PMID:31072929</ref> Although the complex structure was resolved with two molecules of Compound 2 in the hydrophobic binding region, only the proximal inhibitor interacts with the catalytic triad. |
| Line 119: | Line 120: | ||
=== Mutations === | === Mutations === | ||
| - | Missense mutations in LPL or GPIHBP1 | + | Missense mutations in LPL or GPIHBP1 can lead to [https://en.wikipedia.org/wiki/Hypertriglyceridemia hypertriglyceridemia], a condition characterized by high levels of triglycerides in the blood. Two specific point mutations that contribute to this disease through the abolishment of LPL-GPIHBP1 binding are <scene name='87/877554/Met404_cys445/4'>M404R and C445Y</scene>. Because the LPL-GPIHBP1 complex is bound together primarily by hydrophobic interactions, switching a nonpolar [https://en.wikipedia.org/wiki/Methionine methionine] residue to a positively charged [https://en.wikipedia.org/wiki/Arginine arginine] residue, or a nonpolar [https://en.wikipedia.org/wiki/Cysteine cysteine] residue to a polar [https://en.wikipedia.org/wiki/Tyrosine tyrosine] residue will disrupt these hydrophobic interactions and weaken the interface. Also, the much larger side chain of arginine is difficult to fit into the hydrophobic pocket of GPIHBP1. <ref name="Birrane">PMID:30559189</ref> These mutations do not affect LPL’s activity or secretion but play a role its ability to be transported to its site of action in the capillary lumen due to the inability of GPIHBP1 to bind and transport LPL. This leads to an accumulation of unmetabolized lipids but also catalytically active LPL in the interstitial spaces.<ref name="Voss">PMID:21518912</ref> |
| - | Additionally, the carboxylic acid side chain of D201 is critical for coordinating LPL’s calcium ion, and point mutations | + | Additionally, the carboxylic acid side chain of D201 is critical for coordinating LPL’s calcium ion, and the point mutations <scene name='87/877554/Mutation_of_d202_and_d201/12'>D201V and D202E</scene> found in patients with [https://medlineplus.gov/ency/article/000405.htm#:~:text=Chylomicronemia%20syndrome%20is%20a%20disorder,is%20passed%20down%20through%20families. chylomicronemia] have been observed to eliminate LPL secretion and reduce its activity.<ref name="Young">PMID:31269429</ref> Mutating this negatively charged [https://en.wikipedia.org/wiki/Aspartic_acid aspartic acid] into a nonpolar [https://en.wikipedia.org/wiki/Valine valine] residue disrupts the ionic bond between calcium and the aspartate, disturbing the overall calcium binding. A similar mutation has also been observed for D202E. These two mutations, in turn, destabilize LPL folding and thereby prevent its secretion from cells.<ref name="Birrane">PMID:30559189</ref> Knowledge about this enzyme's structure, function, and mutations will help create potential therapeutics to treat chylomicronemia and hypertriglyceridemia, reducing the risk of developing cardiovascular problems.<ref name="Arora">PMID:31072929</ref> |
Current revision
H. sapiens Lipoprotein Lipase in complex with GPIHBP1 and triglyceride metabolism
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Fong LG, Young SG, Beigneux AP, Bensadoun A, Oberer M, Jiang H, Ploug M. GPIHBP1 and Plasma Triglyceride Metabolism. Trends Endocrinol Metab. 2016 Jul;27(7):455-469. doi: 10.1016/j.tem.2016.04.013. , Epub 2016 May 14. PMID:27185325 doi:http://dx.doi.org/10.1016/j.tem.2016.04.013
- ↑ 2.0 2.1 Voss CV, Davies BS, Tat S, Gin P, Fong LG, Pelletier C, Mottler CD, Bensadoun A, Beigneux AP, Young SG. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7980-4. doi:, 10.1073/pnas.1100992108. Epub 2011 Apr 25. PMID:21518912 doi:http://dx.doi.org/10.1073/pnas.1100992108
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ 4.0 4.1 Young SG, Fong LG, Beigneux AP, Allan CM, He C, Jiang H, Nakajima K, Meiyappan M, Birrane G, Ploug M. GPIHBP1 and Lipoprotein Lipase, Partners in Plasma Triglyceride Metabolism. Cell Metab. 2019 Jul 2;30(1):51-65. doi: 10.1016/j.cmet.2019.05.023. PMID:31269429 doi:http://dx.doi.org/10.1016/j.cmet.2019.05.023
- ↑ Olivecrona G. Role of lipoprotein lipase in lipid metabolism. Curr Opin Lipidol. 2016 Jun;27(3):233-41. doi: 10.1097/MOL.0000000000000297. PMID:27031275 doi:http://dx.doi.org/10.1097/MOL.0000000000000297
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Hedstrom L. Serine Protease Mechanism and Specificity. Chemical Reviews 2002 102 (12), 4501-4524 https://doi.org/10.1021/cr000033x
Student Contributors
- Aniyah Coles
- Sarah Maarouf
- Audrey Marjamaa
![Figure 2. A cartoon representation of GPIHBP1's N-terminal intrinsically disordered region (IDR) colored green and an surface representation of LPL with electrostatic coloring [acidic (red), neutral (white), basic (blue)] to show how GPIHBP1's N-terminal domain interacts with LPL's basic patch to stabilize LPL structure and activity.](/wiki/images/thumb/7/72/Electro.jpg/250px-Electro.jpg)